Jason Kim
@jason_z_kim
Followers
2K
Following
245
Media
68
Statuses
300
Postdoctoral researcher at Cornell interested in representation and computation in latent spaces of biological and artificial neural networks.
Joined July 2010
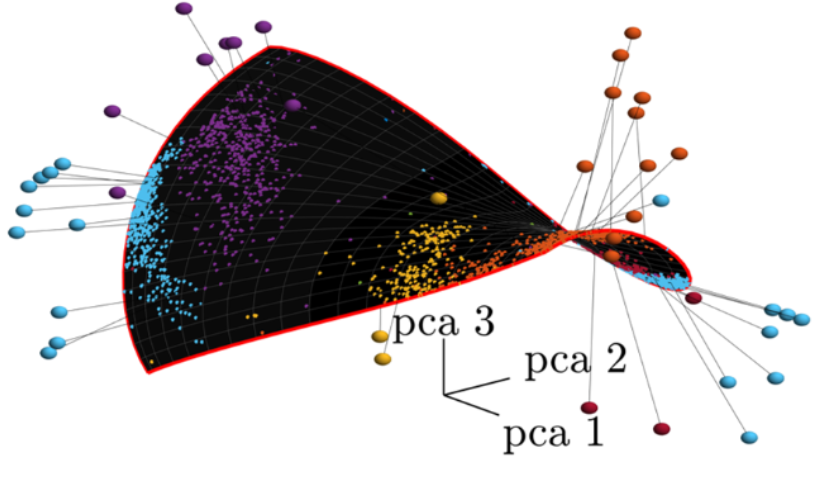
Ever wanted a low-dimensional model of your data that you could be confident would explain data structure and accurately re-embed out-of distribution data, all with minimal distortion of the geometry? Now you can with Γ-VAE! demonstrated on gene data.
1
18
66
RT @LindenParkes: Excited to share the first major piece of work and preprint from my lab! Led by @jason_z_kim! 🥳🎉🤘. .
0
6
0
Working to understand the physics of how the brain works? Interested in understanding how brain function and collective dynamics emerge from neural interactions? Submit an abstract to our focus session "Statistical and Dynamical Physics of the Brain", APS2025 w/@ChrisWLynn!
0
6
30
RT @NatureProtocols: #FeaturedProtocol this week is for a Python-based #software package to apply network control theory to the #humanconne….
0
16
0
RT @LindenParkes: Our protocol paper for NCT is now online at @NatureProtocols!! Check it out here: @jason_z_kim @….
0
25
0
RT @apd_flynn: 🚀 Exciting news alert! 🚀 . Christoph Räth (@DLR_de) and I are thrilled to announce that our minisymposium proposal, 'Dynamic….
0
3
0