alex rubinsteyn
@iskander
Followers
6K
Following
98K
Media
934
Statuses
21K
Genomics + immunology + ML = personalized cancer immunotherapy | https://t.co/nReVwtVHPq | https://t.co/8DWibdfDWa
Durham, NC
Joined June 2007
"Improving long-read somatic structural variant calling with pangenome and de novo personal genome assembly" from @lh3lh3 et al Seemingly big reduction in SV false positives! https://t.co/hZf4Muvdz0
biorxiv.org
Accurate detection of mosaic and somatic structural variants (SVs) provides early diagnostic and therapeutic evidence for cancers. While long-read whole-genome sequencing leads to more accurate SV...
0
1
3
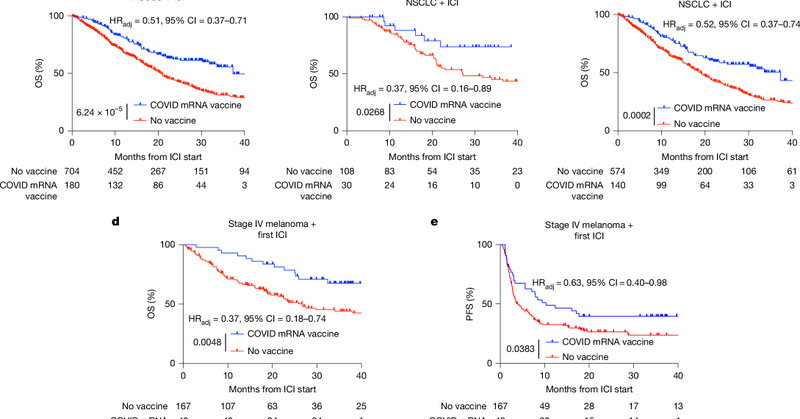
Like, it’s possible that “IFN response to mRNA vaccines” is the dominant reason why aPD1 started having better outcomes after 2020 but, to me, that feels like a stretch. I bet the effect is real but the effect size isn’t.
0
0
2
Maybe a more succinct way to look at it: The curve separation in “SARS-CoV-2 mRNA vaccines sensitize tumours…” is mostly driven by systematic differences in aPD1 outcomes pre-2020 with a sprinkling of comparing effects & health correlates of getting vaccinated during peak Covid
A few different people reached out to see if I believe the results of "SARS-CoV-2 mRNA vaccines sensitize tumours to immune checkpoint blockade" ( https://t.co/qdth5ZDZed) Answer? Kinda. I think the paper conflates three hard to distinguish effects:
2
0
6
“I want to hear the Psoy Korolenko song” -4yo Mission accomplished
0
0
0
Just to clarify: I don't see #1 as a confounder but as The Effect the paper is hoping drives KM split A maybe bigger problem I didn't think enough about that came up in a DM: not getting vaccinated post-2021 is probably correlated with lots of other behavior/health differences
2
0
8
Overall, I'm really puzzled why they didn't look at KM curves with the >100 days sub-group, they pull them out for PD-L1 analysis but for survival. I think all three effects I outlined above are at play here but I'm not sure what their relative contributions are.
1
0
3
The biggest red flag to me is that they're not focusing on the ~1/3rd of their cohort with post-ICI vaccination and comparing it with >100 day before ICI vaccination. That would at alleviate both effects 2/3 & amplify the potential synergy signal between IFN from mRNA and ICI.
1
1
4
The study tries to control for some of these things in a multivariate model but...it's a lot of confounding to try to deconvolve statistically in a small sample.
1
1
3
Patients further back in time were less likely to get aPD1 as a first line therapy, they were more likely to get steroids with aPD1 (making ICI less likely to work), and the management of irAEs hadn't yet been standardized ( https://t.co/ajqGnVoV2e,
https://t.co/EFpw1NOwCP, &c)
1
0
4
(3) the most subtle effect is that Covid vaccine came out in late 2020 / early 2021 but aPD1 came online in the mid-2010s and their cohort starts in 2015. In those ~5 years, hospital practice around checkpoint blockade got way better!
1
2
4
...both the possible anti-cancer adjuvant effects of LNP-mRNA and the protective effects of vaccination against infect (cf https://t.co/afVzEBiF7p -- chemotherapy doesn't totally negate vaccination but drops antibodies by an order of magnitude)
pmc.ncbi.nlm.nih.gov
1
0
3
What about "Patients who received a COVID-19 vaccine within 100 days of chemotherapy...but did not receive ICI had no detectable survival benefit"? Well, one problem with comparing with chemotherapy is that it's typically lymphodepleting and would attenuate...
1
0
3
One reason why this doesn't explain *all* of the effect observed in the paper is that their unvaccinated cohort stretches back to ~2015 and those people weren't dying in eg 2017/2018 of SARS-CoV-2 infection. (We'll get back to this issue of time in the third effect though)
1
0
4
For example, the mortality benefit observed in "Effectiveness of COVID-19 vaccines against severe COVID-19 among patients with cancer in Catalonia, Spain" ( https://t.co/mlie8LfP2E) is similar to the effect observed in the above paper but attributed to protection from SARS-CoV-2
1
0
4
(2) Covid vaccines protect older frail populations from death! It's a shame the other arm is entirely unvaccinated instead of eg vaccine >100 days pre-ICI, comparing vaccinated to unvaccinated cancer patients (mean age 65yo) will always be confounded by the dangers of infection
1
1
5
This effect should also increase with dose of mRNA vaccine (which the paper doesn't see). I think this is a real but likely subtle effect, the smallest of the three which contribute to their separation of survival curves.
1
0
2
(1) adjuvant effect of mRNA vaccination causing additive (if not synergistic) T-cell pressure on cancer cells for patients who get ICI. I think this effect should be most pronounced in the post-ICI vaccination cohort (1/3rd of their cohort, which the paper doesn't split out)
1
1
4
A few different people reached out to see if I believe the results of "SARS-CoV-2 mRNA vaccines sensitize tumours to immune checkpoint blockade" ( https://t.co/qdth5ZDZed) Answer? Kinda. I think the paper conflates three hard to distinguish effects:
nature.com
Nature - mRNA vaccines targeting SARS-CoV-2 also sensitize tumours to immune checkpoint inhibitors.
3
6
17