Clara Groot Crego
@claragrootcrego
Followers
252
Following
532
Media
29
Statuses
207
PhD student in evolutionary biology working on the adaptive radiation of Bromeliads. Also passionate about literature, cinema, music, writing and travel.
Wien, Österreich
Joined February 2018
I am super happy to share our preprint: "Short structural variation fuelled CAM evolution within an explosive bromeliad radiation", taking a closer look at genome evolution in Tillandsia, one of the fastest diversifying clades of the plant kingdom (1/10)
2
10
63
RT @ViePlantGenom: Our newest pre-print links small-scale #StructuralVariation to shifts in #CAM/#C3 #metabolism as key innovation traits d….
0
3
0
Sorryyy, the link was broken! Here the actual link to the preprint:
biorxiv.org
Identifying the drivers of trait evolution and diversification is central to understanding plant diversity and evolution. The subgenus Tillandsia (Bromeliaceae) belongs to one of the fastest radiat...
0
0
3
I thank my wonderful supervisors @ViePlantGenom and @TiBoLeroyInEn, as well as @kheyduk for support, and all other co-authors. This work is dedicated to the late Prof. Christian Lexer, who started this project and passed his love for Bromeliaceae on to me and many others. (10/10)
1
0
7
RT @MedCrisis: By far the most common request I'm emailed is to comment on videos by John Campbell, a popular medical educator on YouTube.….
0
92
0
Super glad to see this work out now in MolEcolRes after such hard times in our research group and in the world! Big congratulations to @GilYardeni for the amazing work towards her PhD and all other co-authors. Proud I could be part of it!
1
2
11
RT @ViePlantGenom: New preprint: an integrated view of metabolism, ecology and transcriptomics in sibling #allopolyploid #Dactylorhiza, ind….
0
10
0
Yesterday I had my second vaccine shot AND this paper from back in my master's came out! What a great day :) Thanks to all the co-authors and especially to my supervisors @RieuxAdrien and Nathalie Becker for everything you taught me while working on this!.
Paper out ! First genome reconstruction of a crop bacterial pathogen from herbarium specimen and its study to enhance our knowledge of its epidemiology & evolution…@claragrootcrego @GagnevinLionel @clbaider @BioAgriOI @Cirad @UPvbmt @UMR_PHIM .
0
3
12
RT @GilYardeni: Hi #Botany2021,.Tomorrow, 11am EST/5pm CEST at Phylogenomics III, I'll talk about some of what we found when comparing our….
0
2
0
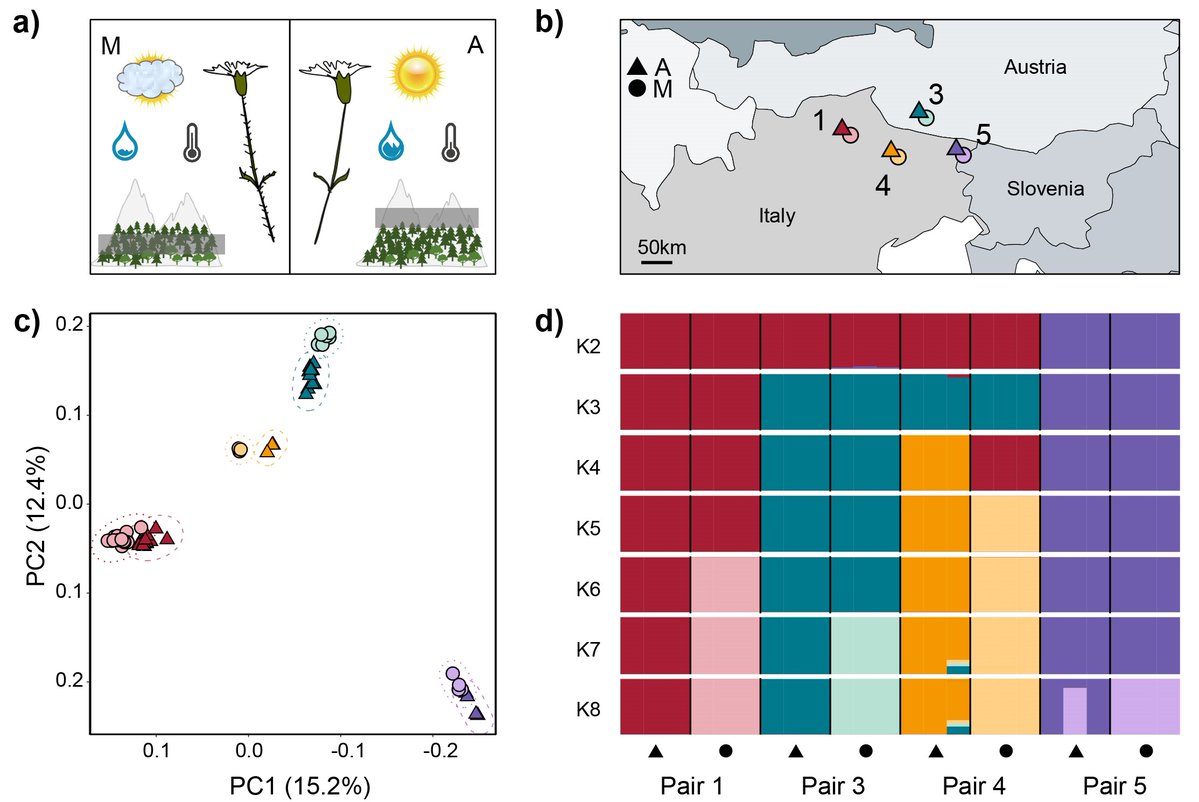
Super proud of my fellow colleague Aglaia Szukala who just got the first part of her PhD work out. She found that the underlying basis of adaptation in repeated ecotype pairs is highly polygenic and achieved through different routes. Really interesting work, check it out!.
A new preprint from our group demonstrates a mostly non-shared genetic basis of parallel #adaptation to different elevations in Heliosperma pusillum, a #Caryophyllaceae. First, coalescent inference confirms parallel polytopic divergence of ecotypes 1/2
0
1
8
RT @GilYardeni: Hi #smbe2021, I'm presenting a poster on our latest preprint (see teaser) in room 9, today & tomorrow 14:00 CET. Let's catc….
0
2
0