Campbell Lab
@camplab1
Followers
104
Following
10
Media
6
Statuses
30
Our group develops and applies novel computational methods to large-scale genomic data in order to gain meaningful insights into disease biology.
Boston University
Joined February 2021
Congrats to Amulya and Charley for their wins at BUMC Evans Day for their talks and posters!
0
0
1
Had a blast tackling the biggest corn maze in New England for our lab fall social!
0
0
1
Recommendations for how to study premalignancy going forward. My contribution was focused on the "organizing principles" of cancer hallmarks. https://t.co/o4tZM1oCBl
nature.com
Nature Reviews Cancer - In this Expert Recommendation, Faupel-Badger and colleagues present a conceptual framework to define precancer and advance our understanding of the earliest changes that...
0
0
1
Excited to publish our collaborative work defining a new standard called "MAMS", which will aid in harmonization and reproducibility of single cell data.
link.springer.com
Genome Biology - Many datasets are being produced by consortia that seek to characterize healthy and disease tissues at single-cell resolution. While biospecimen and experimental information is...
0
1
2
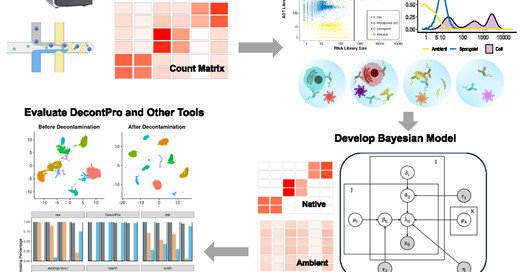
Our publication on how to decontaminate CITE-seq data with DecontPro is out in NAR! Congrats to Yuan Yin for leading a great piece of work!
academic.oup.com
Abstract. Assays such as CITE-seq can measure the abundance of cell surface proteins on individual cells using antibody derived tags (ADTs). However, many
0
0
1
Natasha Gurevich represented our group well at the NCI ITCR annual meeting 2023! She is presenting a novel benchmarking framework for mutational signature algorithms.
0
0
3
**Speaker Spotlight** Joshua D. Campbell Twitter: @camplab1 Website: https://t.co/pMeMdrDnJL
#BioC2023 talk will be on Interactive analysis of single-cell data using flexible workflows with SCTK2.0 Schedule: https://t.co/8Nv8vgAY8F Registration : https://t.co/6eOtFGmtX5
0
4
10
Excited to announce that the last chapter of my PhD thesis has been published! Do I get a bonus for 100% completion? Thanks to Ana Pavel for help getting this across the finish line. Potential roles for miRNAs in COPD and ILD using integrative networks
0
0
0
Here are my 12 guidelines for data exploration and analysis with the right attitude for discovery: 1. You never really finish analyzing a dataset. You just decide to stop and move on at some point, leaving some things undiscovered. 🧵
17
407
1K
Had a great time at the @cziscience #singlecell biology meeting. Met a lot of great people, heard interesting talks, and ate great food!
0
1
3
It is exciting to see that, according to Google Scholar, our tool decontX for decontamination of single-cell data available in @Bioconductor has reach 100 citations! We are happy it has been useful for the community!
link.springer.com
Genome Biology - Droplet-based microfluidic devices have become widely used to perform single-cell RNA sequencing (scRNA-seq). However, ambient RNA present in the cell suspension can be aberrantly...
0
1
3
Our paper for celda is out at NAR Genomics & Bioinformatics! Celda performs co-clustering of genes into modules and cells into subpopulations using single-cell RNA-seq data. https://t.co/zhEZfg08Xz
academic.oup.com
Abstract. Single-cell RNA-seq (scRNA-seq) has emerged as a powerful technique to quantify gene expression in individual cells and to elucidate the molecula
0
10
17
We are excited to join the @cziscience Data Insights program and work on novel methods and software for decontamination of single-cell data!
chanzuckerberg.com
0
0
5
Our lab finally was able to enjoy an "in-person" lab social party which included a BBQ, hike, and a campfire with s'mores.
0
2
8
SAIL engineers helped Junior Faculty Fellow @Josh_D_Campbell & Steering Committee member @wejlab create a tool for comprehensive quality control analysis of single cell #RNA seq data that can be used by anyone online. Learn more on our blog: https://t.co/nt2GebK1ZF
@BUMedicine
0
3
3
The lab is excited to join our collaborators in a #HumanCellAtlas Ancestry Network to characterize prostate cells from different ancestral populations funded by @cziscience. https://t.co/XMW1FHVS76
chanzuckerberg.com
0
3
4
Excited for the publication of musicatk, an R/Bioconductor package for the analysis and visualization of mutational signatures! https://t.co/M24rU9K2xx
0
3
5
Job opening alert! Dr. Jen Beane and Dr. Josh Campbell are seeking a postdoctoral associate - click the link for more information @BUMedicine @camplab1 @Josh_D_Campbell
0
1
1