Replies
Can ML build diverse protein scaffolds around functional motifs? We find equivariant diffusion models and Sequential Monte Carlo may help! Joint work with Jason Yim, and coauthors Doug Tischer,
@ta_broderick
, David Baker,
@BarzilayRegina
and Tommi Jaakkola
8
91
311
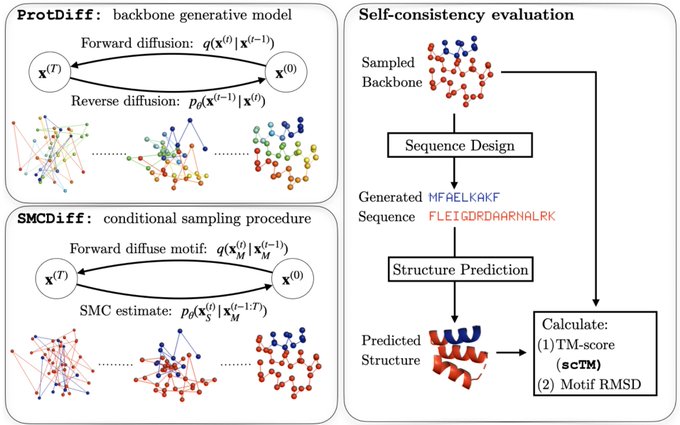
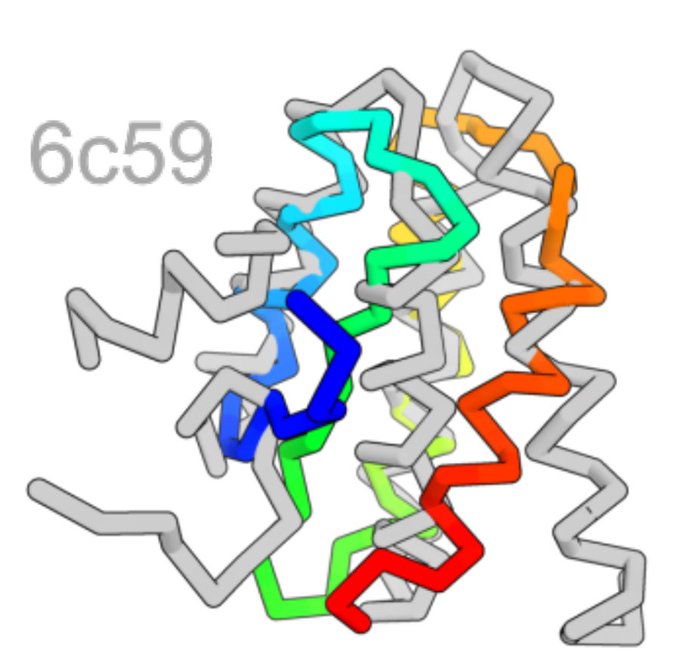
The motif-scaffolding problem is central in protein design: given a target motif (a structural fragment conferring function) construct scaffolds that support it. We propose a generative modeling approach to this problem with two steps.
1
3
17
The first step is modeling a distribution over protein structures. We develop ProtDiff, which tailors equivariant graph neural networks for diffusion probabilistic models (DPMs) on protein backbones. ProtDiff samples diverse topologies, including structures not in the PDB.
1
1
13
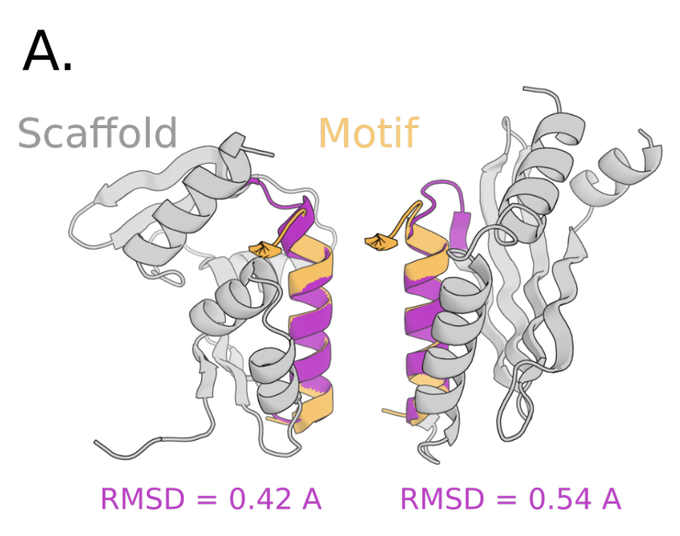
The second step, SMCDiff, samples scaffolds from the conditionals of ProtDiff, given a motif. Naive inpainting fails to generate long scaffolds. So SMCDiff directly targets conditionals of DPMs. It’s the first method to guarantee DPM conditional samples in a large-compute limit.
1
1
12
We model protein backbones only and leverage recent work on fixed-backbone sequence design. We use ProteinMPNN (link) to get sequences and validate them against AlphaFold predictions (without MSA) by TMScore. We call this metric self-consistency TM-Score (scTM).
1
1
10
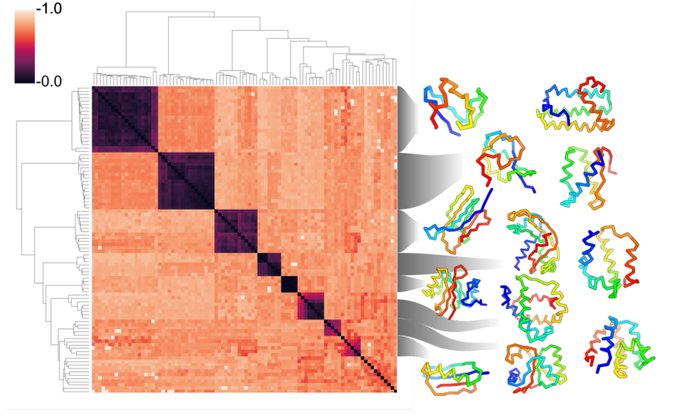
Our samples often agree with AlphaFold predictions (scTM > 0.5) and those that agree exhibit a diverse array of topologies across different lengths.
1
1
9