anna brogan
@apbrogan
Followers
184
Following
295
Media
3
Statuses
64
PhD student into bacteria & coffee. Rudner lab @harvardmed. @penn_state alum.
Boston, MA
Joined May 2019
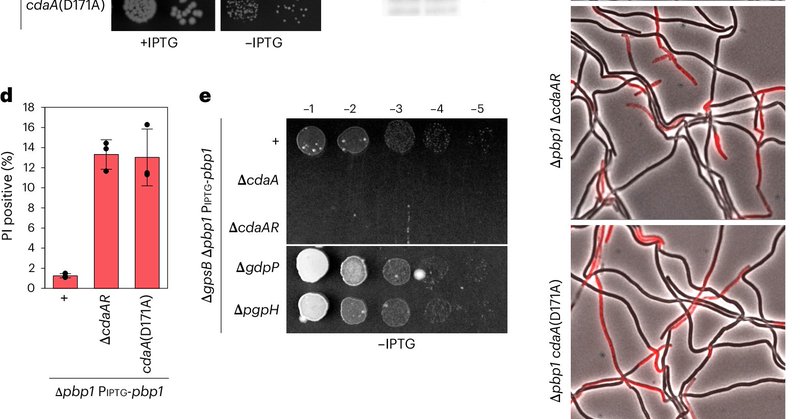
Very happy to share that a large part of my thesis work is out today: B. subtilis uses the second messenger c-di-AMP to modulate its turgor pressure in response to the state of its cell envelope.
nature.com
Nature Microbiology - Brogan et al. uncover a signalling pathway in which levels of the nucleotide second messenger c-di-AMP increase in response to defects in cell wall synthesis. This regulatory...
1
7
39
RT @JorgStulke: The SubtiWiki Paper of the month for June 2025 has been selected! .Congratulations, @apbrogan @ @harvardmed .@NatureMicrobi….
0
3
0
RT @NatureMicrobiol: Out Now! Cyclic-di-AMP modulates cellular turgor in response to defects in bacterial cell wall synthesis https://t.co/….
0
25
0
This was a great collaboration with Dr. Paola Bardetti and Dr. Rico Rojas at NYU. With their help, we provide evidence that c-di-AMP functions to control the cytoplasmic turgor pressure. The c-di-AMP mediated reduction in turgor protects cells from lysis.
0
0
1
In short, there is: the cyclase/regulator complex CdaAR upregulates cyclic-di-AMP in response to defects in cell wall synthesis. Similar to other envelope stress response systems in B. subtills, the cyclase regulator CdaR uses an intrinsically disordered region to sense defects.
1
0
1
c-di-AMP has long been implicated in resistance to cell wall targeting antibiotics. We hypothesized that there could be a regulatory connection between the cell envelope and c-di-AMP levels.
1
0
0
RT @ernstschmidd: Very excited to share that my thesis work is out in Molecular Cell! We trained a Structure and Omics informed Classifier….
cell.com
Schmid and Walter train a classifier that discerns functionally relevant structure predictions in proteome-wide protein-protein interaction (PPI) screens using AlphaFold-Multimer, and they use this...
0
10
0
RT @BostonBacteria: SAVE THE DATE ‼️ 📆 📢.BBM 2025 will be held on June 9-10th at the Harvard Science Center feat. the one and only Dr. Pet….
0
12
0
Angelika Gründling’s work from her sabbatical in the Bernhardt-Rudner labs is out! She came with the goal of finding the missing G+ phosphatidylglycerol phosphate phosphatase and found it with time to spare. I can also highly recommend her as a bay mate!
0
7
29
RT @ThomasMBartlett: The Bartlett Lab is open for business at the Wadsworth Center! We study pathogens with "unusual" shapes (balls & corks….
0
17
0
I had a lovely time getting to hear some fantastic talks and share my own work at the John Innes Centre yesterday! So much exciting microbiology happening.
We’re excited to be kicking off our Early Career Microbiologists Symposium 2024 🌟. Following an introduction from Graham Moore, JIC Director, our morning session is chaired by @SusanSchlimpert and begins with a talk by Anna Brogan @harvardmed
1
5
27
RT @ernstschmidd: Make publication ready figures from your #AlphaFold PAE files. Works on multimer and AlphaFold 3 files. Customize colors….
thecodingbiologist.com
Make high quality publication ready figures from predicted aligned error (PAE) files output by AlphaFold.
0
25
0
RT @ernstschmidd: Interactively browse all the predicted structures here:
predictomes.org
A free interactive resource for viewing predicted AlphaFold multimer structures for protein pairs in biological pathways.
0
2
0
RT @ernstschmidd: Excited to share that our new preprint is out! We used #AlphaFold multimer to screen 7000+ nuclear proteins to find new n….
biorxiv.org
Nucleosomes are the fundamental unit of eukaryotic chromatin. Diverse factors interact with nucleosomes to modulate chromatin architecture and facilitate DNA repair, replication, transcription, and...
0
26
0
another great year for @BostonBacteria! huge shoutout to all of the presenters, the OC, and to @LaurentDubois99 and Molly Sargen for running the whole show (& for incredible crisis management when BBM went dark)
0
1
14
RT @ernstschmidd: Riding the #AlphaFold3 wave with a new analysis tool. It’s super new/unpolished, but would love to get community feedback….
predictomes.org
A free interactive resource for viewing predicted AlphaFold multimer structures for protein pairs in biological pathways.
0
14
0
RT @maxjaderberg: Super excited to be releasing AlphaFold 3 today, developed by @IsomorphicLabs and @GoogleDeepMind: our next generation AI….
0
268
0
RT @EvolvedBiofilm: Bacillus subtilis uses the SigM signaling pathway to prioritize the use of its lipid carrier for cell wall synthesis. @….
0
8
0
Impressive work on a classifier to distinguish likely true AF-multimer predictions from false positives. A bacterial classifier next please!.
My work in Johannes Walter’s lab on developing a classifier to score AF-multimer predictions (alongside a large scale screening effort) is out as a preprint now! @Johanne23743184.
0
0
3