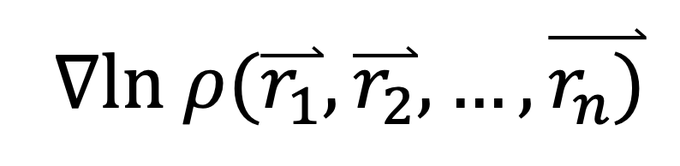So when the diffusion model learns the score it is implicitly learning a free energy and its gradient essentially making it a type of NNP. Now you may object that AF3 doesn't know anything about the forces or the energies as it is trained on the PDB so how can it possible be…
1
2
11
Replies
Ok so the new AlphaFold model relies in large part on a "relatively standard diffusion approach" turns out you can think of this as just a special case of a neural network potential, it just uses experimental data not quantum chemistry to train on. 1/n
6
42
305
First off a neural network potential is just a very flexible function with a ton of parameters that takes in positions and outputs energies/forces. More detail in this thread:
1
3
13
In contrast diffusion models learn the "score" which is the gradient of the log of a probability distribution. For AF3 this is just the probability of the atoms having a particular position. In stat mech log probs are free energies or potentials of mean force and their grads are…
1
3
16
The answer is that it actually does know something about the forces it knows that the average forces on the proteins in the PDB are 0. If they weren't zero then they wouldn't be in equilibrium and you couldn't crystallise them and obtain their structure with XRD. If the average…
1
1
9
What about away from the minima? Well it just assumes the force/score points back to the minima linearly proportional to the distance away from it. That what's 'denoising' is you're just telling it the force points in the opposite direction to the noise on the positions.
1
2
7
If you trained on the true equilibrium distribution structures extracted from a simulation you get the true forces (if your noise is sufficiently low) This paper first showed this and we have validated for a simple system that it is precise.
1
4
26
This approximation of linear forces back to minima has a long history in physics its called the harmonic approximation and physicist famously use it everywhere. It results in a Gaussian probability distribution and a nice smooth surface to optimise on. This paper outlines this…
1
2
12
Another trick diffusion models use? They start with a high noise level and then gradually reduce it to refine the distribution. This is essentially thermal annealing another tool from stat mech.
1
1
12
And a final trick? They use Langevin dynamics for inference. A standard molecular simulation algorithm invented by a Physicist. When you run Langevin dynamics you get Boltzmann probabilities. i,e,. exponential of the free energy. So we end up back with original probabilities.
1
1
10
This means that Google's claim that they have "surpassed physics based tools" is kind of strange. In fact there is a ton of physics baked into it how diffusion models work!
1
4
31
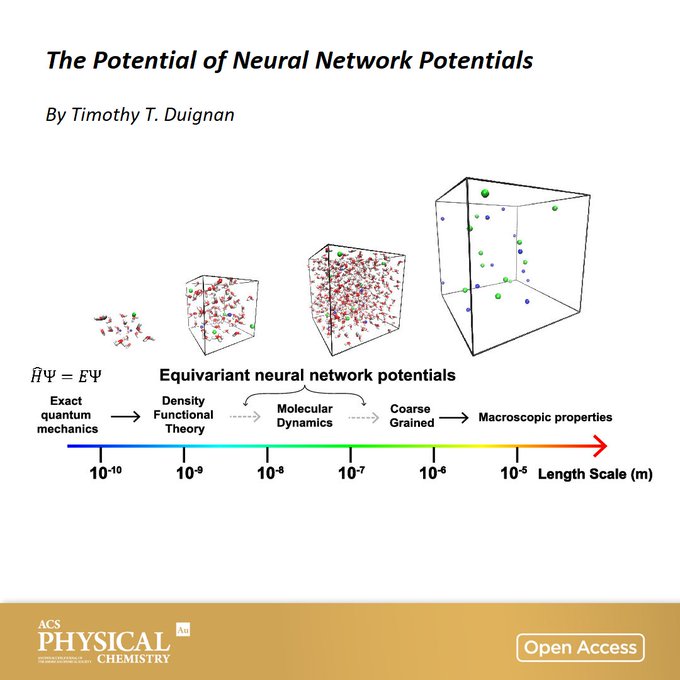
More info in this thread and paper:
The Potential of
#Neural
Network Potentials
A perspective from Timothy Duignan
@TimothyDuignan
@Griffith_Uni
🔓 Open access in ACS Physical Chemistry Au 👉
1
35
192
1
3
10