Thomas Walter
@ThomasW04410315
Followers
246
Following
224
Media
15
Statuses
140
Full Professor (AI) at Mines ParisTech. Director of the Centre for Computational Biology, Mines ParisTech. Researcher in Bioimage Informatics.
Paris, France
Joined October 2019
We hope this contributes to open & reproducible #ComputationalPathology. Big thanks to @guillaumebalezo, Albert Pla Planas and Etienne Decenciere. Great collaboration between @Sanofi and @Mines_Paris, supported by @ANRT. #DigitalPathology #FoundationModels #ViT #HuggingFace.
0
0
1
🧪 Try it yourself!.We've released an interactive demo on Hugging Face 🤖.👉 👨💻 Code: 📂 Data:
zenodo.org
Source: Mapillary 4DCity URL: https://4dcity.org/imgupload/1656605088.084.jpg Original Image URL: https://scontent-muc2-1.xx.fbcdn.net/m1/v/t6/An_vp0A00iAvwI6vytfUcAQ0SFLLth9JjQsT6uWENDZ71-mfZ0A8V6...
1
0
2
💡 The core idea of MIPHEI-ViT was to use a ViT foundation encoder (H-optimus-0) for this dense prediction task, thus leveraging the power of pathology foundation models for cross-modality prediction.
1
0
1
🔬 H&E stained tissue slides are cheap, and available for huge retrospective cohorts. mIF (and in particular Orion) are much more informative on particular cell types, but not available for large-scale cohorts. 💡 MIPHEI-ViT bridges the gap, learning to predict mIF from H&E.
1
0
1
🚨 New preprint + open-source release!.We're excited to share MIPHEI-ViT — a model that predicts multiplex immunofluorescence (mIF) from H&E-stained histology images using Vision Transformer (ViT) foundation models. 📄
arxiv.org
Histopathological analysis is a cornerstone of cancer diagnosis, with Hematoxylin and Eosin (H&E) staining routinely acquired for every patient to visualize cell morphology and tissue...
1
4
9
👩💻 Please checkout our code:
github.com
Contribute to 15bonte/cell_cycle_classification development by creating an account on GitHub.
1
0
1
🔬 We’re also releasing a massive new dataset — over 600,000 annotated nucleus images — now freely available on the BioImage Archive. 🔗
ebi.ac.uk
BioStudies – one package for all the data supporting a study
1
0
0
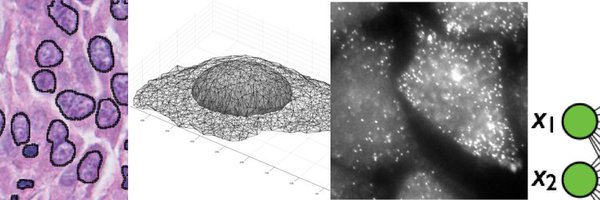
🧠 We introduce Cell-Cycle Variational Auto-Encoders (CC-VAE) — a deep learning framework to robustly and consistently predict cell cycle phase from microscopy images.
1
0
0
🎉 We're excited to share our latest work: "A Deep Learning approach for time-consistent cell cycle phase prediction from microscopy data", now available on bioRxiv!. 📰
biorxiv.org
The cell cycle consists of four phases and impacts most cellular processes. In imaging assays, the cycle phase can be identified using dedicated cell-cycle markers. However, such markers occupy...
1
1
4
Hopefully a useful read for anyone tackling spatial transcriptomics and trying to get down to the single-cell level. 3/3. @institut_curie @Inserm @Mines_Paris . #PRAIRIE.
0
0
1
Huge thanks to @LGaspardBoulinc & @Luca_Gortana for this amazing work and @fmgcavalli and Emmanuel Barillot for this wonderful collaboration at the U1331 - Computational Oncology 2/3.
1
0
1
Excited to share our new review in Nature Reviews Genetics!. "Cell-type deconvolution methods for spatial transcriptomics".🔗 📖 Free access: 1/3.
1
3
6
RT @aipulserx: Can AI predict complex multiplex immunofluorescence markers from standard H&E images, potentially revolutionizing cancer tis….
0
5
0
- RNA2seg package: - Annotated datasets: - Preprint: Looking forward to feedback from the community!.
1
0
3
Accurate cell segmentation is critical in spatial transcriptomics but often challenged by poor staining and complex tissues. RNA2seg addresses this by integrating all available data types—membrane, nuclear staining, and RNA positions.
1
0
4
RNA2Seg is a deep learning model trained on over 4 million cells across 7 organs, integrating RNA point clouds and multiple stainings for robust and accurate cell segmentation.
1
0
1
🎉 Happy to share our new preprint!. Congratulations to @faraone92406241 and @AliceBlondel4 for their outstanding work on RNA2seg – a generalist model for cell segmentation in image-based spatial transcriptomics. And thanks Florian Müller for the nice collaboration!.
1
2
4
Join us to innovate and lead in digital health!. Apply now! #JobOffer #Bioinformatics #AI #DigitalHealth. More information and application procedure:.
0
0
1
Exciting Opportunity at the Center for Computational Biology!. We're hiring a Junior Professor Chair in Bioinformatics/Computational Biology: "Artificial Intelligence for Digital Health." Competitive startup package & a path to full professorship in 3-6 years.
2
7
9