Phil b. Pope
@ThePopeLab
Followers
2K
Following
3K
Media
66
Statuses
2K
Playing with all sorts of weird and wonderful microbiomes since 2014 @CMR_QUT and @uniNMBU. Ambassador for @ISME_microbes
Australia
Joined August 2010
Some news to share😀 I will be relocating to The Centre for Microbiome Research (@CMR_QUT and @QUT) in Feb 2024 to launch a @arc_gov_au #FutureFellowship with Gene Tyson and @S_J_Mc that brings omics-guided high throughput cultivation to the rumen microbiome. New bugs = new clues.
16
5
115
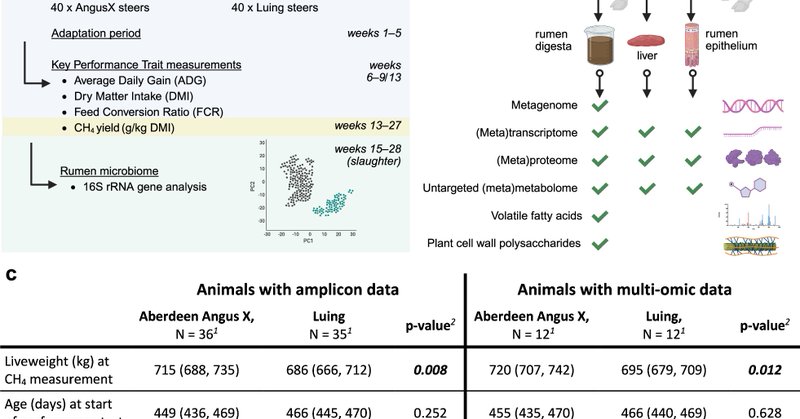
Back in 1962 microscopy was used to pen a hypothesis about eukaryote-microbe relationships. ~65 years later we have coupled the latest advancements in long read MetaG, MetaP and EUK SAGs to help explain what this means for wider rumen microbiome function
nature.com
Nature Communications - Here, the authors reveal that protozoal communities shape rumen microbiome structure, offering fresh insights into how these complex communities coordinate essential...
0
4
19
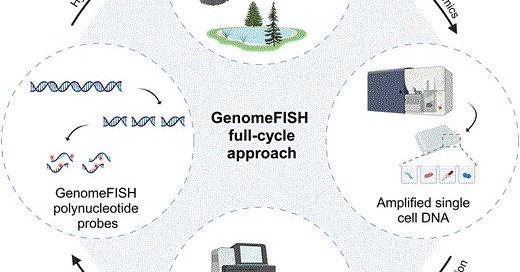
RT @jcamthrash: GenomeFISH: genome-based fluorescence in situ hybridisation for strain-level visualisation of microbial communities https:/….
academic.oup.com
Abstract. Fluorescence in situ hybridization (FISH) is a powerful tool for visualizing the spatial organization of microbial communities. However, traditio
0
18
0
RT @LabMizrahi: Our new preprint, "Spatial constraints drive amylosome-mediated resistant starch degradation by Ruminococcus bromii in the….
0
2
0
The Centre for Microbiome Research @CMR_QUT is looking for a postdoc dedicated to advancing our understanding towards the microbiological, biochemical and genetic processes that cause methanogenesis in the cow rumen. More info 👉
2
17
36
RT @JensWalter15: Very much to our surprise, a strain of Limosilactobacillus reuteri drives severe pathology in a mouse model of multiple s….
0
10
0
We are looking for PhD students to join us at @CMR_QUT. A range of projects on offer covering gut microbiome in animal health and nutrition, microbial responses to climate change and microbial methane mediation. More info found here 👉
0
4
11
RT @LabMizrahi: Fun to share our data and hear about the amazing science and developments in metaproteomics at the 6th International Metapr….
0
3
0
RT @TRHvidsten: It is still possible to apply for PhD scholarships within computational host/microbiome genomics (hologenomics) at the Norw….
0
7
0
RT @magnusarntzen: We look forward to an exciting talk by Emma Timmins-Schiffman on "Uncovering microbial mysteries: novel peptide sequenci….
0
3
0
RT @microbetracker: Our study on drug-gut microbiome interactions is out! 🦠💊 @NatureMicrobiol. We show that the #Parkinson's drug entacapo….
0
24
0
RT @CMR_QUT: A huge congratulations to A/Profs @S_J_Mc and @ThePopeLab on their induction into the ARC's College of Experts! 🎉 We are confi….
0
1
0
RT @CMR_QUT: Congrats to @ThePopeLab on the recent publication in Communications Biology, which demonstrates the need for high-resolution m….
nature.com
Communications Biology - A multi-omic approach enables the reconstruction of microbial metabolic dynamics in the salmon gut in response to feed and feed supplements, outlining novel and potentially...
0
1
0
RT @greeninglab: So excited to see this out! Aerated environments can be major sources of methane. This is due to the activities of aerotol….
biorxiv.org
Methanogenesis is classically thought to be limited to strictly anoxic environments. While oxygenated oceans are a known methane source, it is argued that methanogenesis is driven by methylphosphon...
0
22
0
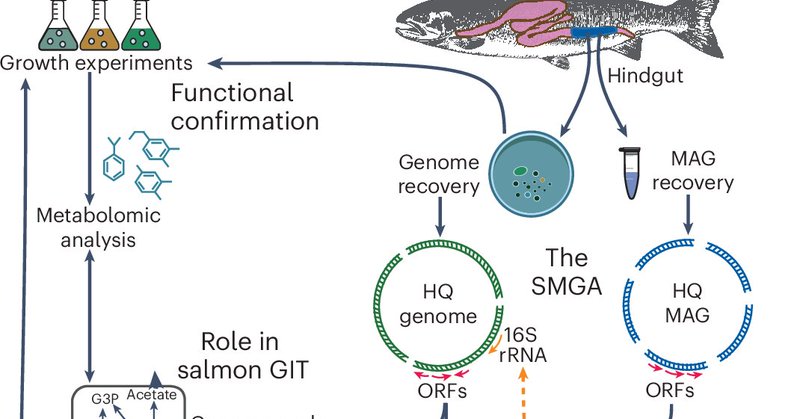
RT @NatureMicrobiol: NEW RESOURCE: Using shotgun metagenomics, cultivation and metabolic modelling, the Salmon Microbial Genome Atlas will….
nature.com
Nature Microbiology - Using shotgun metagenomics, cultivation and metabolic modelling, the authors construct the Salmon Microbial Genome Atlas as a resource for future studies on sustainable...
0
11
0
RT @CMR_QUT: Check out the latest article from CMR’s @ThePopeLab, published in @NatureMicrobiol. The study explores how bacteria found in t….
0
6
0
4 years of collaboration has poured into this resource! Massive thanks to @SabinaLLaRosa and @quark_buzo_vera for driving this and of course all our partners coming from @UniNMBU and abroad @srsandve @TRHvidsten @MortenLimborg @ines_thiele @Stebe69.
0
1
7
A fishy tale about the the Salmon Microbial Genome Atlas is now out in @NatureMicrobiol. We hope this culture and genome biobank provides a functional resource to the community and supports the next generation of salmon gut microbiome projects.
nature.com
Nature Microbiology - Using shotgun metagenomics, cultivation and metabolic modelling, the authors construct the Salmon Microbial Genome Atlas as a resource for future studies on sustainable...
2
22
72
Anyone working with metaproteomics and looking for a conference 500m away from a ski resort join us for IMS6 in Norway, January 13-15, 2025. Registrations and abstract submissions are open now!.#metaproteomics #metagenomics #microbiome . Sign up here:.
0
3
13
RT @TRHvidsten: We are looking for two PhD students to join our @UniNMBU team to explore host-microbe interactions using genomics. The pos….
0
18
0
RT @HoloGen_MSCA: 🚀 Exciting news! The new @MSCActions Doctoral Network @HoloGen_MSCA is kicking off on Sept 9th! Led by Prof. @TRHvidsten….
0
7
0