Stephane Redon
@StephaneRedon
Followers
2K
Following
649
Media
834
Statuses
2K
Co-founder and CEO at OneAngstrom | I post about molecular design and #SAMSON, the integrative molecular design platform | https://t.co/c5qr0P6BZa
Joined February 2012
🎉🎉🎉 Today, we have a huge announcement to make: SAMSON is now free for non-commercial use! This includes all extensions for docking, simulating, animating, scripting, and much, much more. Precisely, we are making the entire SAMSON molecular design platform - SAMSON + every
16
189
783
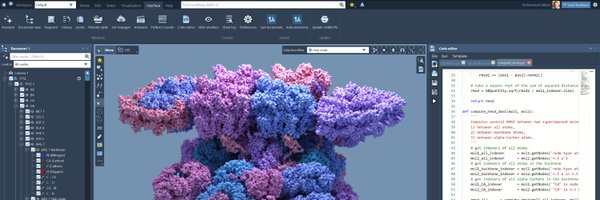
Once paths are found, examine results in detail. Each path's conformation and energies are essential for understanding protein dynamics.
1
0
0
Configure search parameters, including seed usage, ARAP iterations, and minimization steps, for optimal path discovery outcomes.
1
0
0
A precise sampling box is crucial. Start with a box that encloses all atoms, then adjust dimensions to bias motion as needed.
1
0
0
Define active ARAP atoms to guide protein motion effectively. SAMSON allows easy selection and visualization of these key atoms.
1
0
0
Select start and goal conformations from your active document. These serve as the seeds for finding the transition path.
1
0
0
Critical step: Set the interaction model to Universal Force Field (UFF) and choose FIRE as your state updater to ensure accurate calculations.
1
0
0
Launch the Protein Path Finder app and explore its dual tabs: 'Settings' to configure parameters and 'Results' to collect your discoveries.
1
0
0
In SAMSON, start by preparing your protein model. Check for alternate locations, remove ligands and solvents, and add hydrogens if necessary.
1
0
0
First, ensure you have the necessary extensions. The Protein Path Finder app and FIRE state updater are essential for this task.
1
0
0
Learn how to efficiently determine paths between protein conformations with SAMSON's Protein Path Finder. This thread will guide you through the process!
1
0
4
Options for initial box fitting include specifying box lengths or solute-box distance. This flexibility ensures you can optimize for both performance and accuracy in your simulations.
0
0
0
When preparing your system, you can choose your unit cell shape and size tailored to your needs. The goal is to fit your model as efficiently as possible without unnecessary complexity.
1
0
0
Using a rhombic dodecahedron saves about 29% of CPU time when simulating due to its smaller volume, compared to a cube with the same image distance. Ideal for flexible molecules in solvent.
1
0
0
The rhombic dodecahedron and truncated octahedron unit cells are closer to a sphere compared to a cube. This makes them more efficient for spherical macromolecule simulations, saving resources. 🧬
1
0
0
Did you know that GROMACS supports multiple unit cell shapes for space-filling? This includes cubic, orthorhombic, triclinic, rhombic dodecahedron, and truncated octahedron.
1
0
0
Explore how choosing the right unit cell shape can optimize your molecular simulations. This thread dives into the various shapes supported by GROMACS and their benefits. #Thread
1
0
1
Molecular dynamics of a coupler that locks two shafts together. It's easier to connect short pieces together than make and place one really long shaft.
189
213
5K
🔍 Dive into the full documentation for a more comprehensive understanding of each attribute and how they revolutionize molecular design in SAMSON Connect.
1
0
0