Prathith Bhargav | ಪ್ರಥಿತ ಭಾರ್ಗವ
@PrathithB
Followers
224
Following
3K
Media
2
Statuses
100
BS-MS from @ChemistryIISERP @IISERPune '24. 🖥Biophysics and Biochemistry | Machine Learning | Epidemiology | Science Education | Swimming | Carnatic Music
Pune, India
Joined May 2021
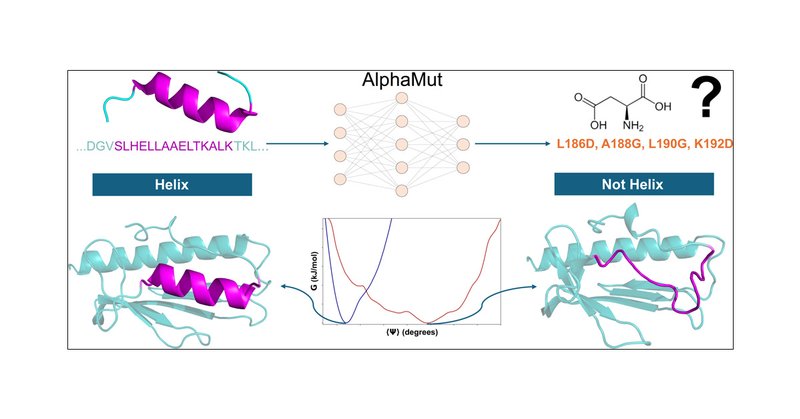
I am thrilled to announce that my MS Thesis work with @ArnabPune is now published in @JCIM_JCTC! Check out the paper in the link below. @ChemistryIISERP @IISERPune.
pubs.acs.org
Helices are important secondary structural motifs within proteins and are pivotal in numerous physiological processes. While amino acids (AA) such as alanine and leucine are known to promote helix...
I'm excited to announce that my MS project work @IISERPune is now communicated as a preprint! Check out the quoted tweet to find how we used reinforcement learning to disrupt alpha helices in proteins. Many thanks to my supervisor @ArnabPune for all the guidance and support! 1/n.
8
5
59
RT @ArnabPune: Happy to share our latest publication with @PrathithB on protein structure modification using a fixed set of mutations --Al….
pubs.acs.org
Helices are important secondary structural motifs within proteins and are pivotal in numerous physiological processes. While amino acids (AA) such as alanine and leucine are known to promote helix...
0
4
0
RT @workshopmlsb: Starting in 5! Join us for an exciting day of invited talks, posters, and a panel with our invited speakers. Schedule: h….
0
6
0
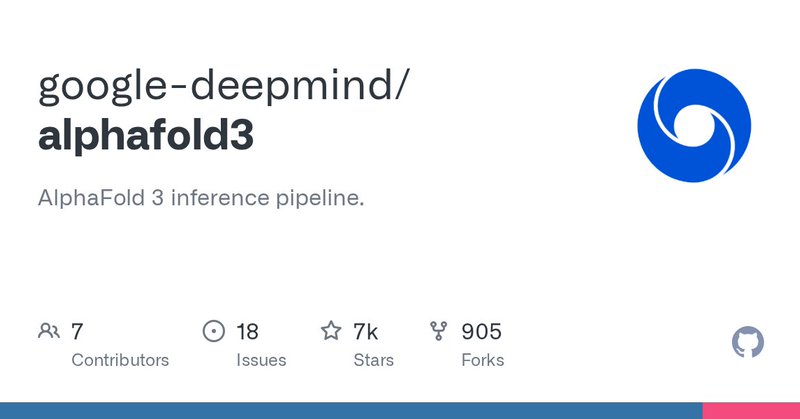
RT @pushmeet: The #AlphaFold 3 model code and weights are now available for academic use. We @GoogleDeepMind are excited to see how the res….
github.com
AlphaFold 3 inference pipeline. Contribute to google-deepmind/alphafold3 development by creating an account on GitHub.
0
263
0
RT @roybikirna: Delighted to present our work on memory dependent friction @IISERPune. Many thanks to my supervisor @ArnabPune and Hridya V….
0
2
0
Chemistry Nobel for Protein Structure Prediction and Protein Design!!!.
The Royal Swedish Academy of Sciences has decided to award the Nobel Prize in Chemistry 2024 with one half to David Baker “for computational protein design”, and the other half jointly to Demis Hassabis and John M. Jumper “for protein structure prediction”. Read more:
0
0
3
@BiologyAIDaily @biorxiv_bioinfo If you're interested in breaking helices from proteins of your choice, check out this colab notebook: colab works okay till 250 aa length proteins) 4/n.
colab.research.google.com
Run, share, and edit Python notebooks
0
0
3
@BiologyAIDaily With @biorxiv_bioinfo partnering with ScienceCast, you can also listen to an AI generated summary of the work. 3/n
1
0
3
@BiologyAIDaily has compiled a nice summary of the work! 2/n
AlphaMut: a deep reinforcement learning model to suggest helix-disrupting mutations. - AlphaMut is a novel reinforcement learning model designed to identify mutations that disrupt protein helical structures, providing a powerful tool for protein engineering and functional
1
0
1
I'm excited to announce that my MS project work @IISERPune is now communicated as a preprint! Check out the quoted tweet to find how we used reinforcement learning to disrupt alpha helices in proteins. Many thanks to my supervisor @ArnabPune for all the guidance and support! 1/n.
AlphaMut: a deep reinforcement learning model to suggest helix-disrupting mutations #biorxiv_bioinfo.
8
4
75
The moment we were all waiting for.
Announcing AlphaFold 3: our state-of-the-art AI model for predicting the structure and interactions of all life’s molecules. 🧬. Here’s how we built it with @IsomorphicLabs and what it means for biology. 🧵
0
0
5
Next up in the #Byelingual series, I just realised I just cannot remember some words in Hindi. What's the word for pillow? Dimbu(oh no, that's Kannada), Ushi (oh no, that's Marathi). Proceeds to say Ushi and finds out the other person is Marathi. Felt good, after all.
0
0
14