Zuobai Zhang
@Oxer22
Followers
696
Following
1K
Media
29
Statuses
123
Research intern @nvidia; Ph.D. student at @Mila_Quebec. Interested in deep generative model, drug discovery and protein science.
Montreal, Quebec
Joined September 2021
🚀 Excited to share Proteina, our protein structure generative model! Trained on 21M protein structures with scaled-up model size, with state-of-the-art designability and diversity. Huge thanks to the incredible NVIDIA GenAI team for an amazing internship experience! 🌟 #GenAI.
📢📢 "Proteina: Scaling Flow-based Protein Structure Generative Models". #ICLR2025 (Oral Presentation). 🔥 Project page: 📜 Paper: 🛠️ Code and weights: 🧵Details in thread. (1/n)
2
6
42
Interesting analysis from Pascal! Combining different modalities for protein representation learning will be the future🚀🚀🚀.
Have we hit a "scaling wall" for protein language models? 🤔 Our latest ProteinGym v1.3 release suggests that for zero-shot fitness prediction, simply making pLMs bigger isn't better beyond 1-4B parameters. The winning strategy? Combining MSAs & structure in multimodal models!.
0
0
3
RT @HannesStaerk: At #ICLR2025 in Singapore now. Looking for people to chat with :) . Also presenting ProtComposer:
0
7
0
RT @DidiKieran: Excited to present my first paper officially as a PhD student now as an ICLR Oral this week! Super fun work with the GenAIR….
0
15
0
RT @karsten_kreis: 🔥 I'm at ICLR'25 in Singapore this week - happy to chat!. 📜 With wonderful co-authors, I'm co-presenting 4 main conferen….
0
7
0
RT @ArashVahdat: My GTC talk highlighting some of the Gen AI for science projects from my team at NVIDIA and the lessons we've learned alon….
0
18
0
RT @karsten_kreis: 🔥Large-scale protein structure generation? Flow Matching? Fold class guidance? Big transformers? State-of-the-art benchm….
0
8
0
RT @karsten_kreis: 🔥Excited to present *Proteina* next week (accepted as Oral to ICLR'25) in the ML Protein Engineering Seminar Series. Hea….
0
9
0
We’re giving a tutorial on protein design at #AAAI2025! Come and join us if you’re interested.
🤔Excited about Protein + AI? Join our "AI for Protein Design" tutorial at #AAAI2025, with gorgeous @Oxer22 @divnori @WengongJin @tangjianpku and Jiwoong Park. 🧬✨. 📅 8:30–12:30 (EST), Feb 26 .📍Room 117, Philadelphia Convention Center, Philadelphia.👉
1
1
19
RT @XinyuYuan402: 🚀scCello (NeurIPS2024 Spotlight) is now open-source! .We’re excited to release the code and data for scCello, a cell-onto….
0
10
0
RT @karsten_kreis: @ArashVahdat and I will chat about opportunities in NVIDIA's fundamental generative AI research team. 🔥 Protein and mol….
0
7
0
I’ll present our work tomorrow afternoon at East Exhibition Hall #1211. Come and have a chat if you’re interested in protein design and representation learning! #NeurIPS2024.
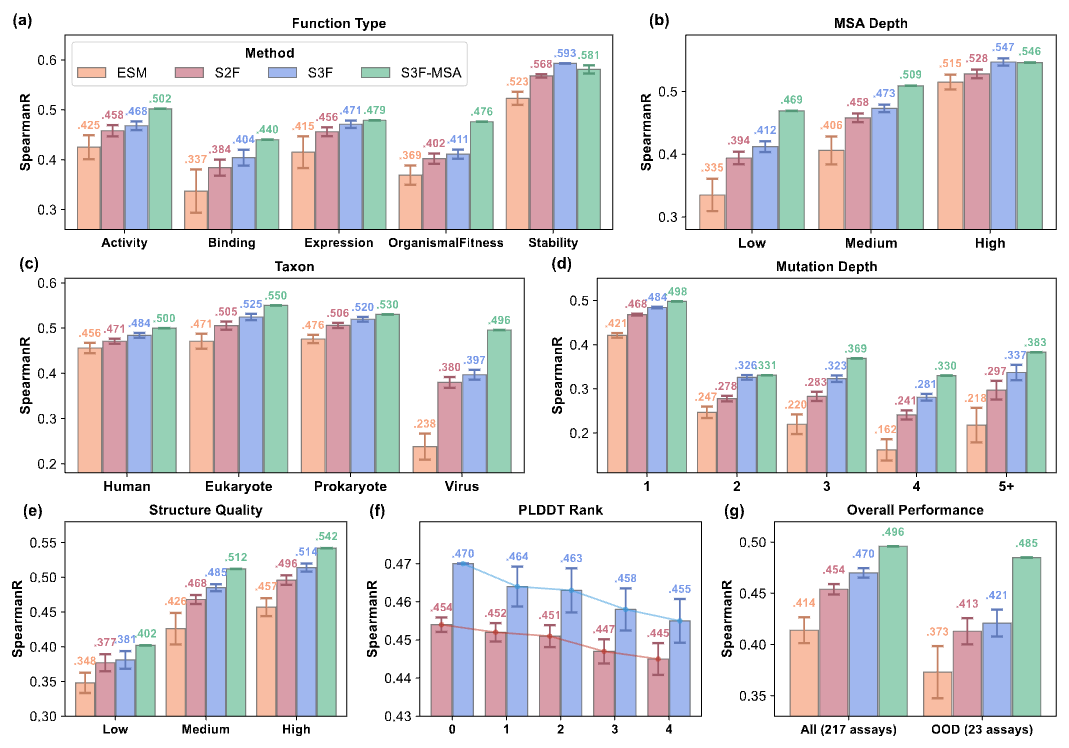
🤔 How does structural information help in protein fitness prediction? . 🚀 Excited to share our work S3F (Sequence-Structure-Surface Fitness) model at #NeurIPS2024, a multimodal model for zero-shot protein fitness prediction! 🧬 #AIforScience.Paper: 1/🧵
0
1
13
🙏 A huge thank you to my incredible collaborators @NotinPascal, Yining, Aurelie, @vijiltw, @deboramarks, @payel791 and my advisor @tangjianpku for making this work possible!.
1
0
2
Curious to learn more? Explore our code here:.👉 We can’t wait to discuss this at #NeurIPS2024 in Vancouver! 🌟 🇨🇦.6/🧵.
1
0
2