M3s (B. Hatzikirou)
@M3sBiomath
Followers
424
Following
2K
Media
85
Statuses
635
Associate professor in mathematical biology @KhalifaUni ° Group leader @tu_dresden/RT is not endorsement
Abu Dhabi, UAE
Joined March 2015
RT @keenanisalive: Here's a nice "proof without words":. The sum of the squares of several positive values can never be bigger than the squ….
0
577
0
RT @Jun_kiim: Just out in Science (2025)—a landmark study by @Dr_CJackson et al. identifies a previously uncharacterized immune cell popula….
0
124
0
RT @mcapellanus: For those interested, some details of the forthcoming book "Mathematical Oncology", Chaplain & Preziosi:. 🧵 .
0
93
0
RT @guillelorenzogz: 📢📜 New #mathonco preprint out ! Mathematical models can forecast tumor growth and treatment response, but robust strat….
arxiv.org
Despite advances in methods to interrogate tumor biology, the observational and population-based approach of classical cancer research and clinical oncology does not enable anticipation of tumor...
0
3
0
RT @Kos_Marinakis: Επειδή έχουν ακουστεί ονόματα σταθμών και δημοσιογράφων που καμία σχέση δεν έχουν με την υπόθεση, να ξεκαθαρίσω πως το β….
0
2K
0
RT @fabian_theis: 1/🚀 Excited to share RegVelo, our new computational model combining RNA velocity with gene regulatory network (GRN) dynam….
0
98
0
RT @mkjolly15: Excited about this collaborative work in @iScience_CP on comparing the Notch signaling models with least microenviroometal u….
0
3
0
I am very excited that our collaboration of.@arnabbarua10 and @mkjolly15 team (Aditi Pujar,.@UshasiRoy2, @divyoj and Partha Dey) is finally published. Thread follows /1
cell.com
Molecular network; Biocomputational method; Flux data; In silico biology; Biological constraints
6
9
25
RT @sisyga91: Thrilled to see our latest research featured in the MathOnco blog! Thanks for the chance to share our work with such a wide a….
journals.plos.org
Author summary Intratumor heterogeneity presents a significant barrier to effective cancer therapy. This heterogeneity stems from the evolution of cancer cells and their capability for phenotypic...
0
3
0
Our new study on understanding tumor heterogeneity with @sisyga91 , @Andreas52436912 ,Harish Jain and Marcus Kellner published in
journals.plos.org
Author summary Intratumor heterogeneity presents a significant barrier to effective cancer therapy. This heterogeneity stems from the evolution of cancer cells and their capability for phenotypic...
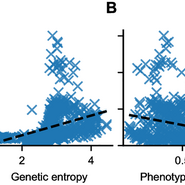
Tumor heterogeneity is a major obstacle to effective cancer treatment. It is caused by multiple factors, including irreversible mutations and reversible phenotypic changes. But how can we model the interplay between both processes and what's their effect on cancer treatment? 1/
0
0
10
RT @pejmanshojam31: Had a fantastic time at the final conference of my PhD journey in Toledo. Grateful to the organizers of @ecmtb2024 for….
0
2
0
RT @pejmanshojam31: Ismaila Mohammad from @KhalifaUni team give his talk at the session „model reduction and identifiability“. @ecmtb2024….
0
1
0
RT @pejmanshojam31: Attended to the first session „mechanistic learning in mathematical oncology“ organised by @AlvaroKohn. Babis @M3sBioma….
0
2
0
RT @pejmanshojam31: Excited to share my latest research on predicting glioma recurrence! Using an in-silico model, we found that macrophage….
0
2
0
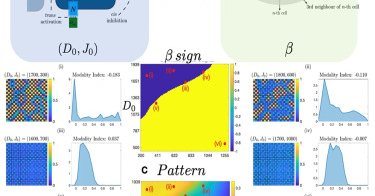
@arnabbarua10 A trade-off between synchronization time and sensing radius, showing optimal conditions for information transfer.
0
0
2