Linzy Elton, PhD
@LinzyElton
Followers
774
Following
1K
Media
32
Statuses
2K
#GlobalHealth researcher #AMR #sequencing, football fan & lover of all dogs that have ever existed. Postdoc for @pandoraIDNet @UCL_CCM, PI @AmrCovid (she/her)
Joined May 2015
Don't forget, there's a plain language summary of our recently pubished paper utilising #metagenomics to identify #pathogens and #AMR in veterinary hospital environments on my blog:
linzyelton628749128.wordpress.com
Also available as a podcast here. This is a plain language summary of a paper we (myself and colleagues across UCL and the Royal Veterinary College (RVC) wrote, which describes how we used sequenci…
0
0
0
Published! Our paper utilising #metagenomics to look at #pathogens and #AMR in the veterinary environment, and how it relates to clinical infections:
microbiologyresearch.org
0
3
6
We held our Lab Olympics today, congratulations to all competitors, especially team 'Number one' for coming first!
1
2
11
The team at @ZymoResearch have written a blog about the use of environmental #metagenomics in two of our recent papers, highlighting its use for #AMR #surveillance across #OneHealth settings
bioscience.co.uk
0
1
1
If you're working on #genomics #surveillance #OneHealth and #AMR, consider publishing your research in the Antibiotics special issue! Link below 👇
📚Special Issue: Genomic Surveillance of Antimicrobial Resistance (AMR) 😊 Edited by Dr. Linzy Elton 📆Submission Deadline is 30 September 2025. 🔗 https://t.co/u4UyOlCBbn
0
0
6
In a collaboration between @HerpeZ, @UCL_CCM and @NHSBartsHealth, researcher Rosalind @roro_micro has spent 3 weeks in Zambia helping to develop @nanopore #metagnomics for non-malarial febrile illness diagnosis from blood samples! @JohnTembo1982 @kathrynharris26 @LinzyElton
0
5
8
In our final video on #clinicaltrials, @LinzyElton explains what a site coordinator does when they visit each of the labs! Coordinators work closely with the lab teams to help make sure that all work is being done according to the trial guidelines, making the data robust.
0
4
6
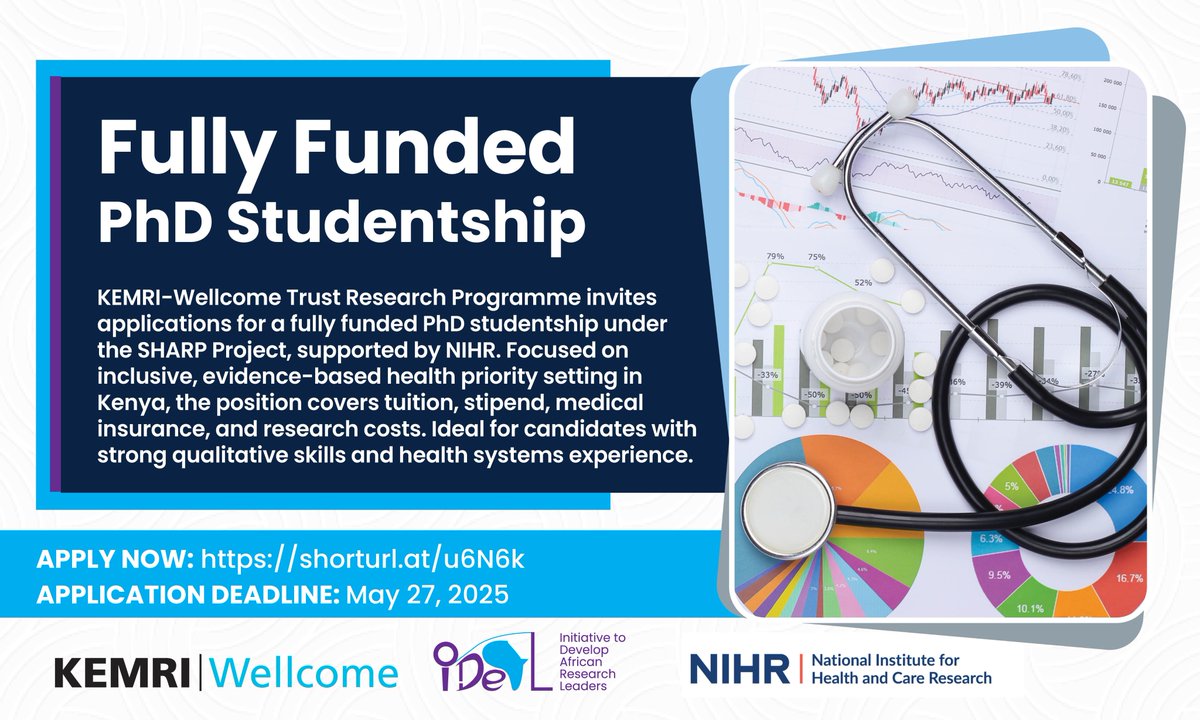
Apply now for a fully funded PhD with KEMRI-Wellcome Trust, supported by NIHR! Conduct your research at a world-class research programme, with complete support for tuition, stipend, and research costs. Click here to apply: https://t.co/m8PwBre4rg Deadline: May 27, 2025
2
63
92
Next up in our series of videos on #clinicaltrials, @LinzyElton explains what a day in the life of our laboratory scientists looks like! We work with sites all over the world and each trial is different, but with many hundreds of #tuberculosis patients enrolled, it's always busy!
0
3
6
We're part of many #tuberculosis #ClinicalTrial studies, and site coordinator @LinzyElton has made a series of videos to try and dispel the myths about clinical trials! First up, a look at OptiRimox; why (and how) it's run, and why trials are so important!
0
3
5
Alright so here we go! We conducted a clinical trial in children under the age of 1 who were HIV positive and had severe pneumonia. The mortality rate in these children has been unacceptably high even with empirical antibiotic treatment.
26
67
253
Msc and PhD opportunities with ZNPHI sponsored by the World Bank ACDC project.
1
29
28
(1/2) Are you interested in @nanopore sequencing? Want to do whole genome sequencing? Our postdoc @linzyElton has published a @YouTube tutorial video for the Rapid Barcoding kit (RBK114) to help you get started!
1
3
3
(1/2) Are you interested in @nanopore sequencing? Want to sequence amplicons or do #metagenomics? @linzyElton at our partner site @UCL_CCM @ucl has published a @YouTube tutorial video for the Rapid PCR Barcoding kit to help you get started:
1
4
4
My blog covering the plain language summary of our recent paper is now out! Learn how we used metagenomics to identify #AMR genes and clinically relevant bacterial species in a companion animal hospital in London!
linzyelton628749128.wordpress.com
Also available as a podcast here. This is a plain language summary of a paper we (myself and colleagues across UCL and the Royal Veterinary College (RVC) wrote, which describes how we used sequenci…
0
1
2
(5/5) Discussion: We identified AMR genes, plasmids and species of relevance to human and animal medicine. When considering IPC measures, adherence to, and frequency of, cleaning schedules, alongside potentially more comprehensive disinfection of animal-handling areas is vital
0
0
0
(4/5) Results: Common AMR genes identified were aph, sul, blaCARB, tet and blaTEM. In clinical isolates, many isolates had β-lactam resistance. Areas with high animal handling had the highest number of AMR genes. 24 Gm+ and 4 enterobacterial plasmids were identified
1
0
0
(3/5) Methods: Samples were collected in a vet hospital in London. Metagenomic DNA was sequenced using @nanopore. The sequence data were analysed for AMR genes, plasmids and pathogen species. These were compared to phenotypic speciation and ASTs of bacteria isolates from patients
1
0
1
(2/5) Intro: There are currently no standardised guidelines for genomic surveillance of #OneHealth #AMR. We utilised #metagenomics to identify AMR genes present in a companion animal hospital and compare with phenotypic results from clinical specimens from the same hospital
1
0
0