Eric Sun
@EricDSun
Followers
957
Following
123
Media
13
Statuses
64
Machine learning for aging, spatial/single-cell omics, immunology || Incoming Assistant Professor @MIT | PhD @Stanford
Stanford, CA
Joined October 2021
I’m very excited to be joining MIT Biological Engineering @MITdeptofBE as an Assistant Professor and the Ragon Institute @ragoninstitute as a member in January 2026! I will be recruiting students and postdocs (see more info below).
30
36
709
#Agents4Science received over 300 submissions and accepted 48 papers. Check them out here: https://t.co/mpp2rxEOC7 Many of these AI agent-written papers are quite interesting! The online conference will be on Oct 22; you can register for free at https://t.co/dD3sNnLBHT
6
39
177
I would like to extend a big thank you to my amazing co-advisers @james_y_zou & @BrunetLab and to my many fantastic mentors, colleagues, and collaborators!
1
0
9
My lab will build AI/ML tools and computational methods to model the biology of aging & rejuvenation with a particular focus on spatial and single-cell omics and immune cell interactions.
1
0
13
Spatial transcriptomic clocks reveal cell proximity effects in brain ageing https://t.co/7rEYok8Tct
nature.com
Nature - A spatially resolved single-cell transcriptomics map of the mouse brain at different ages reveals signatures of ageing, rejuvenation and disease, including ageing effects associated with T...
1
5
29
How do cellular neighbors shape the aging brain? Knight Initiative researchers Anne Brunet @BrunetLab and James Zou @james_y_zou merge spatial transcriptomics and AI to uncover how local cellular interactions drive brain aging and resilience. Learn more: https://t.co/Ciu1lGbeQ4
1
4
17
A big thank you to Anne Brunet (@BrunetLab) and James Zou (@james_y_zou) for their fantastic mentorship and to all co-authors for their valuable contributions -- this work would not have been possible without them!!!
2
0
4
Of course more work is needed to determine how much each of these mediators contributes to these proximity effects and whether T cells or NSCs can be targeted to improve brain function.
1
1
13
Taking these predictions to the lab, we were able to confirm spatial colocalization of markers for each of these mediating pathways!
1
0
0
And we identified lipid metabolism (in nearby cells) and exosomes/growth factors (in NSCs) as potential mediators for the NSC pro-rejuvenating effect.
1
1
4
Using TISSUE, we identified interferon signaling as a potential mediator of the T cell pro-aging effect.
1
1
1
What are the potential mediators driving the pro-aging effect of T cells or the pro-rejuvenating effect of NSCs? We turned to TISSUE, a method we previously developed to impute spatial gene expression and perform statistical tests:
nature.com
Nature Methods - Transcript Imputation with Spatial Single-cell Uncertainty Estimation (TISSUE) offers a general framework for estimating uncertainty for spatial gene expression predictions,...
1
0
0
Excitingly, some interventions like exercise can modulate the cell proximity effects in a rejuvenating directions (for example reducing the T cell pro-aging effect)!
1
2
2
We also found the pro-aging effect of T cells and the pro-rejuvenating effect of NSCs across multiple external datasets and these results were robust to different modeling approaches (see Extended Data of the paper for details).
1
0
1
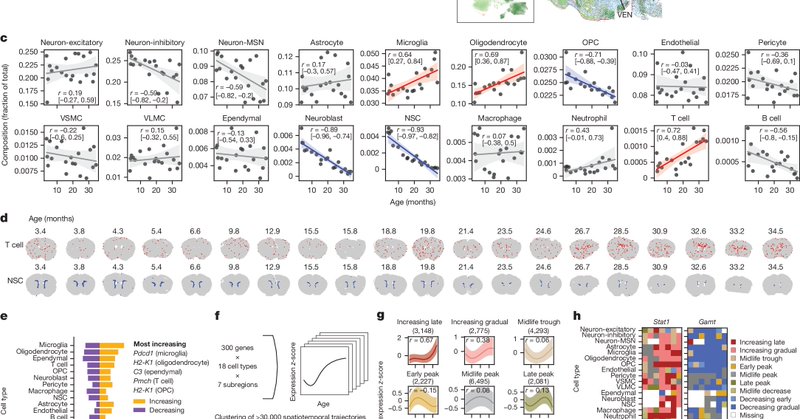
We extended the proximity effect analysis to the whole neighborhood level using graph neural networks (GNNs), which also allowed us to perform "in silico" perturbations and examine their effects on neighborhood aging--again finding the pro-aging T cells and pro-rejuvenating NSCs.
1
0
1
Summarizing these effects, we found that T cells had the most pro-aging proximity effect and neural stem cells (NSCs) had the most pro-rejuvenating proximity effect on average.
1
1
2
Now, using the spatial aging clocks as a tool for discovery, we wondered whether certain cell types could influence the aging of nearby cells? We computed "cell proximity effects" with the spatial aging clocks for all pairings of cell types.
1
1
0
While we were working on this project, several exciting spatial datasets on brain disease or perturbations became available. Applying our clocks to these datasets showed accelerated aging for certain cell types (particularly glia)! See our paper for more results.
1
0
0
We also generated MERFISH data for young and old mice undergoing rejuvenating interventions (exercise and partial reprogramming). Spatial aging clocks revealed that these interventions impact different cell types and different brain regions!
1
0
0