Ashesh Ghosh
@AsheshGhosh1
Followers
217
Following
2K
Media
23
Statuses
69
postdoc @UCBerkeley prev: postdoc @Stanford, Grad @UofIllinois interests: Eq/Non-eq Stat Mech| Soft Matter, Glass & Polymer Physics| Biophysics| Fluid dyn
Mountain View, CA
Joined March 2020
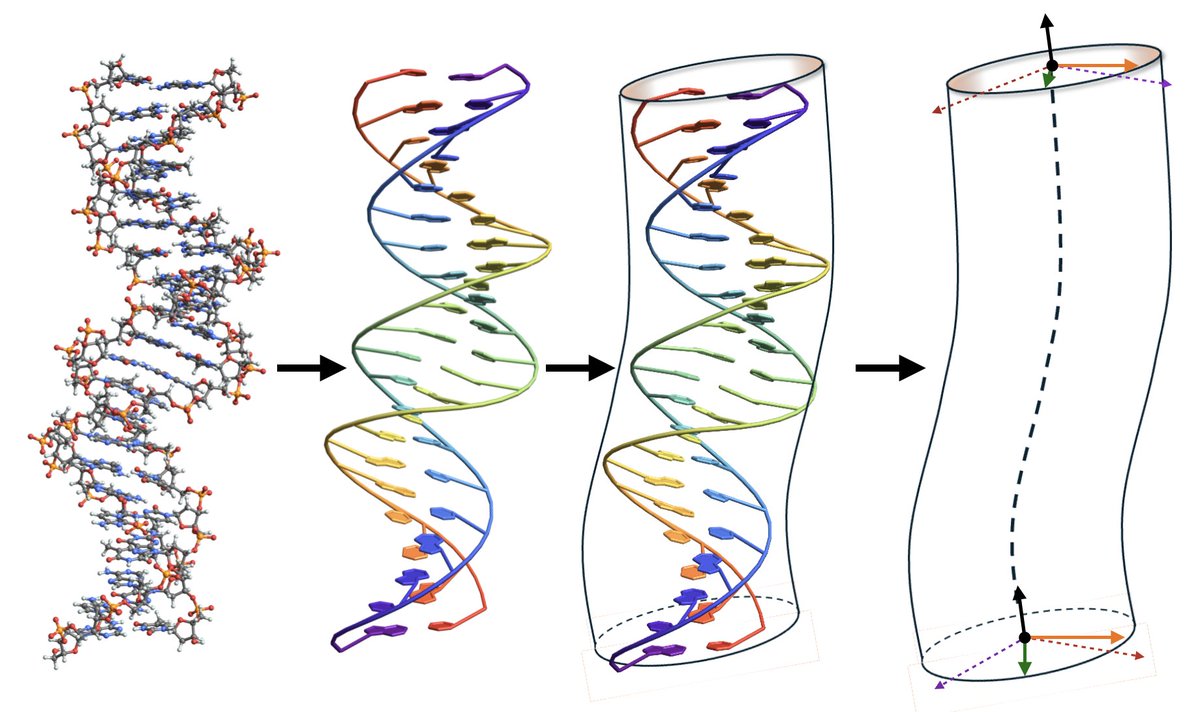
(1/2) The simplest models of dsDNA maps the double helix to a continuous rod/cylinder with twists and bends along the polymer. The groove asymmetry of DNA results in coupled twist-bend modes. Here we solve for the exact chain statistics and . .
When is DNA like an asymmetric top? Check out @AsheshGhosh1's study on the twisted worm-like chain, a good model for DNA mechanics, whose Green's function is solved in a basis of Wigner D-functions, the eigenfunctions of the asymmetric top @UCB_Chemistry
1
0
10
Excited to share this latest work on a crucial aspect of endosome tethering and fusion. The work shows how small scale changes can sometimes drive large scale coordinated transformations. Have a look at this work @
Ashesh Ghosh and Andy Spakowitz published “Local changes in protein filament properties drive large-scale membrane transformations involved in.endosome tethering and fusion” in Soft Matter. Excellent work, Ashesh!
2
1
25
RT @ASpakowitz: Sayantan Dutta, Ashesh Ghosh, and Andy Spakowitz published “Effect of local active fluctuations on structure and dynamics o….
0
3
0
(1) Can we say if active forces are at play by looking at polymer structure and/or single-/multi-point trajectories of polymers? .(2) If yes, which locations along the chain experience activity? How are they correlated?.@SayantanD2 answers these here 👇.
How does spatial and temporal correlation of active forces impact structure and dynamics of biopolymers? We adapt the physics of flexible polymers and demonstrate using an e.g. of point fluctuation in this work with @AsheshGhosh1 and @ASpakowitz now online in @softmatter (1/2).
1
0
5
Large scale biophysical changes are often driven by orchestrated events at much finer length and timescales. With @ASpakowitz we show how local filament properties drive large-scale membrane transformations. Preprint at
1
2
23
RT @SayantanD2: How do we construct the connectivity between loci of chromosome marked with the same color? We propose an algorithm founded….
0
4
0
🙂👇.
Accounting for many-body correlation is crucial to understand slow dynamics of supercooled liquids. This article elucidates the role of deGennes narrowing in collective Debye-Waller factors and glassy dynamics of model hard sphere fluids. @AsheshGhosh1.
4
0
15