OpenProtein.AI
@openprotein
Followers
234
Following
7
Media
7
Statuses
31
Joined June 2023
Boltz-1 & Boltz-2 now live via GUI & APIs! Predict protein, protein–RNA/DNA/ligand structures with confidence scores & binding affinity metrics for virtual screening. Compare finetuned models in the new overview page to find your best performer fast. https://t.co/wNZPlwLksh
0
1
3
Product update: Indel Analysis lets you score insertions/deletions across your sequence using PoET-2. You can now also compare multiple 3D structures in Mol* to evaluate design alternatives. Sign up now: https://t.co/wNZPlwKMCJ
0
2
4
Why does no one in AI protein engineering work on indels? We’re solving this at https://t.co/61ZJL2kIf4. Check out our upcoming indel design tooI! 🤩 1/4
2
11
69
Product update: PoET-2 now supports structure inputs for enhanced prediction and design via Python APIs. Check out our new inverse folding tutorial to see it in action. 🔗 https://t.co/0mflT7X6ai Sign up for https://t.co/NGI2kaXi7Z:
openprotein.ai
Join the revolution in protein research with early access to our cutting-edge Open Protein AI platform. Sign up now to explore the future of protein analysis and discovery.
0
4
9
🧬 Protein Revolution: The Tiny Model Making a Massive Impact! PoET-2 is changing the game in computational protein design, slashing experimental data needs by 30x! 🚀 learn more: https://t.co/zNbMYILkJb
#ProteinDesign #BiotechInnovation #AIRevolution
0
3
10
This is just the beginning of what's possible with AI that truly understands the molecular machinery of life. Join us in transforming protein engineering:
openprotein.ai
Join the revolution in protein research with early access to our cutting-edge Open Protein AI platform. Sign up now to explore the future of protein analysis and discovery.
0
0
0
Ready to try it yourself? PoET-2 is available now on https://t.co/NGI2kaXi7Z: - Free academic access - Python client & APIs - Web interface
1
0
0
Want to see the technical details? Read our white paper:
openprotein.ai
PoET-2 is a next generation protein language model that transforms our ability to engineer proteins by learning from nature’s design principles. Through its unique multimodal architecture and...
1
0
2
The implications are enormous for: - Drug discovery - Enzyme engineering - Protein therapeutics - And much more
1
0
1
This means PoET-2 doesn't just memorize - it learns fundamental principles of how proteins work, enabling accurate zero-shot variant effect prediction and highly data efficient property learning.
1
0
0
How does it work? PoET-2's tiered attention mechanism processes large protein families with order equivariance and long context lengths, letting it learn from evolutionary examples at inference time.
1
0
0
In real-world testing, PoET-2 can: - Design proteins with multiple simultaneous constraints - Learn from just dozens of examples - Make accurate predictions for challenging proteins - Run fast inference on standard hardware
1
0
1
PoET-2 introduces a powerful prompt grammar for controlled protein generation - enabling everything from inverse folding to motif scaffolding in a single model.
1
0
0
The results are remarkable: - 500x more compute efficient than contemporary models - 30x less experimental data needed for protein optimization - Improved on structure understanding - Handles insertions and deletions naturally
1
0
0
Key breakthrough: PoET-2's multimodal architecture learns to reason about sequences, structures, and evolutionary relationships simultaneously through in-context learning.
1
0
1
Most protein language models rely on massive scale - up to 100B parameters - to memorize sequences from nature. PoET-2 takes a fundamentally different approach, learning the grammar of protein evolution.
1
0
0
🧬 Announcing PoET-2: A breakthrough protein language model that achieves trillion-parameter performance with just 182M parameters, transforming our ability to understand proteins.
1
9
21
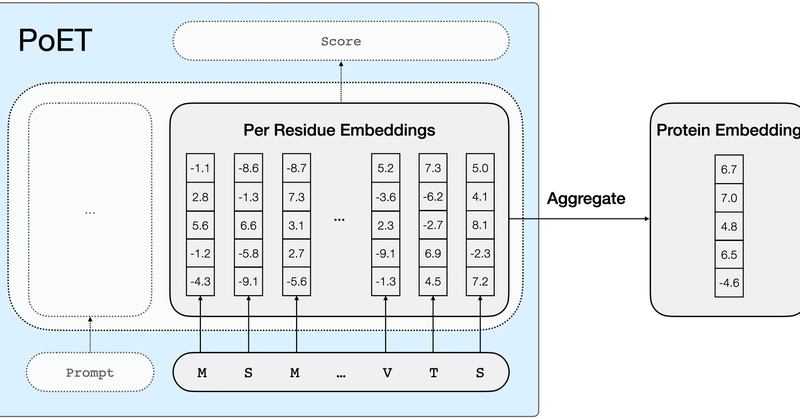
New blog post! PoET, our generative protein language model, is also super powerful for fine-tuning. PoET outperforms previous methods for supervised variant effect prediction even with 15x less data.
openprotein.ai
PoET, our generative protein language model, is a powerful foundation model for supervised learning on proteins. Remarkably, PoET-based models significantly outperform previous methods for predicting...
0
3
8
Engineering protein complexes or multimeric proteins is challenging! Our platform lets you model critical interactions between subunits easily. See our latest walkthrough for an example of how to design optimized variants with cross-chain mutations.
0
0
4