NIHR GHRU AMR
@GHRUamr
Followers
675
Following
72
Media
52
Statuses
174
National Institute for Health Research Global Health Research Unit on Genomic Surveillance of AMR
Joined October 2018
The @NIHRglobal @GHRUamr focuses on sustainable Genomics for national #AMR surveillance in Colombia, India, Nigeria, and the Philippines and local / global genomic context for ongoing surveillance of major #AMR pathogens. Here we detail our work with a series of preprints (1/10)
1
23
33
RT @TheCGPS: A fantastic achievement by our @GHRUamr partner @SomosAGROSAVIA in becoming the first GFGP certified organisation in the Ameri….
0
9
0
RT @HarryHarste: Huge congratulations to @SomosAGROSAVIA in becoming GFGP gold certified! Testament to the hard work of @MARCELA59964548 @e….
0
6
0
Very enjoyable 14th TGIBF (Thank Goodness It's Bioinformatics Friday) session today - sharing genomic AMR surveillance results across all the GHRU units and learning together. Today there were 20 participants from @RITMPH @SomosAGROSAVIA @CentralResearc2 @UniIbadan @TheCGPS
0
5
21
Enjoyed our 12th TGIBF (Thank Goodness It's Bioinformatics Friday) meetup convened by @bioinformAnt.Discussion on best practice lab and bioinformatics for @nanopore sequencing of bacteria + SARS-CoV-2. Presentations from Sam Dougan (@TheCGPS) & @varunshamanna (@CentralResearc2)
0
6
12
RT @NIHRglobal: The rise of antimicrobial resistance (AMR) is a global healthcare issues. Prof David Aanensen of the NIHR Research Unit on….
0
2
0
RT @ghruamrcol: 👸/🍄Antimicrobial resistance research should have a strategic agenda and the application of cutting-edge technologies for th….
0
4
0
RT @iruka_okeke: Many thanks to all that made this visit such a pleasure. Grateful for support we receive from @NIHR & @FlemingFund, and o….
0
2
0
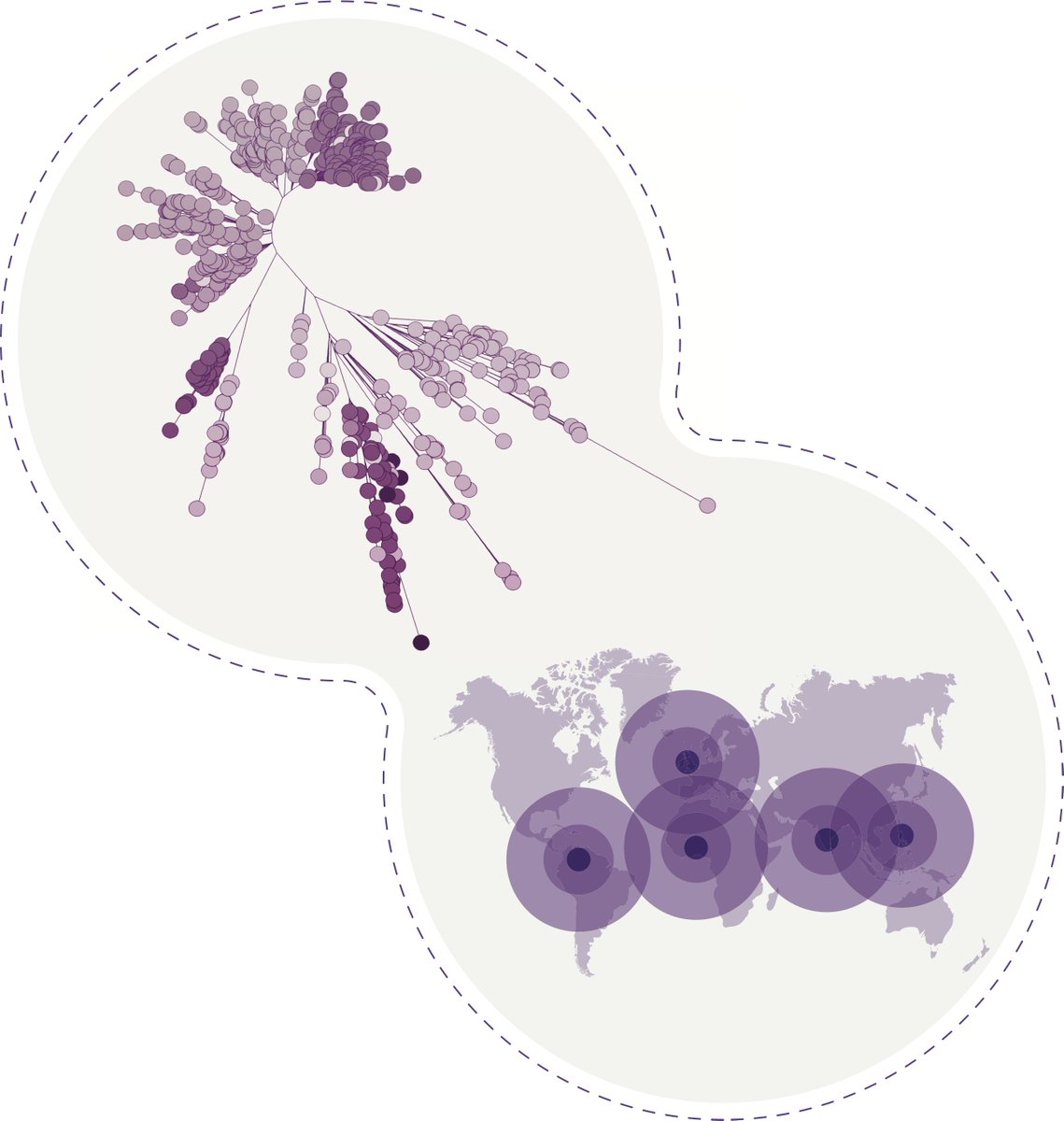
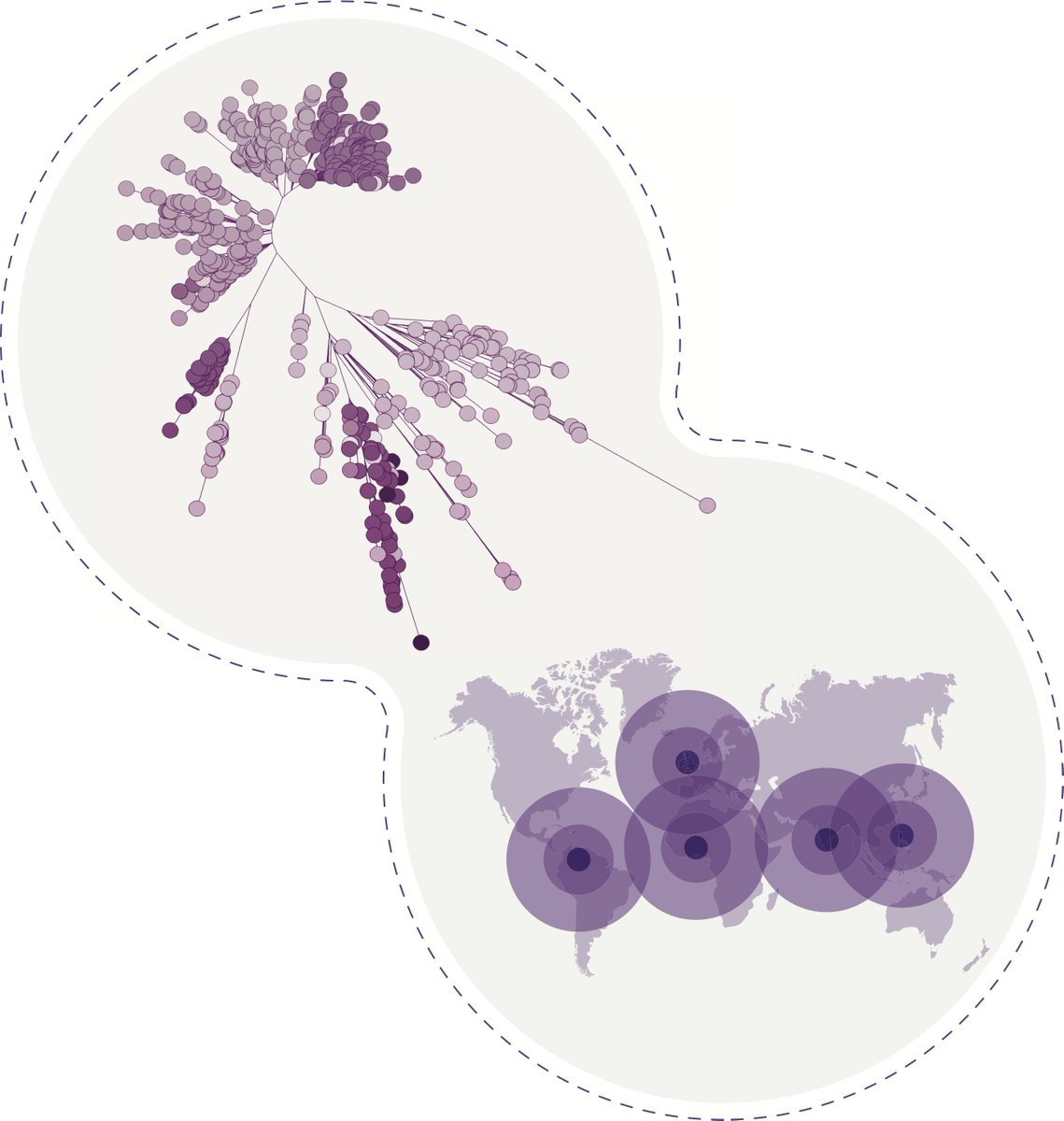
Here we detail the global landscape of Klebsiella genomics and provide an ongoing resource for surveillance through @pathogenwatch We analyse 1636 genomes sequenced by our consortium together with 16,537 public genomes (all with geospatial data).(10/10)
0
1
2
Nigeria: @UniIbadan uncover known+novel lineages of K. pneumoniae including MDR ESBL-producers & rarely carbapenemase-producing isolates enabling the identification of putative outbreak clusters in hospitals. @Ayorinde_Afo @anderson_ose @iruka_okeke (8/10)
1
1
1
India: Here, @CrlKims highlight the presence of high-risk international Klebsiella clones and identify a persistent outbreak of carbapenem and cephalosporin resistant ST231 isolates in one hospital. @Geetha66538200 @varunshamanna. (7/10)
2
1
1
Colombia: .@SomosAGROSAVIA. describe how Klebsiella pneumoniae clonal expansions of CG258 carry KPC-3 and NDM-1 whilst other high-risk clones such as CG147, CG307, and CG152 carry diverse carbapenemases .@ghruamrcol @pilarxavier @INSColombia .(6/10)
1
1
2
Training: Here we detail our Train-the-Trainer approach to apply effective methods for training others in bioinformatics and laboratory techniques required for genomic surveillance of #AMR. @MonicaAbrudan @alicepn @dusanka @eventsWCS. (5/10)
1
2
5
Financial Capacity Building: International efforts to standardise management of grants are covered by Good Financial Grant Practice (GFGP). Here we describe the use of GFGP to increase institutional capacity .@HarryHarste @gennykiff @iruka_okeke. (4/10)
1
1
1
Bioinformatics and data flow: Here we detail bottlenecks in bioinformatics deployment and routes to overcoming these by including standardised workflows across all laboratories. @Ayorinde_Afo @varunshamanna @turrisita @MonicaAbrudan @bioinformAnt .(3/10)
1
2
4
Laboratory Implementation : An interactive approach to implement genomic sequencing across sites via lab assessment, assembly, optimisation, and re-assessment to deliver surveillance capacity. @cake_ray @Geetha66538200 @anderson_ose @ghruamrcol.(2/10)
1
1
1
The @NIHRglobal @GHRUamr. focuses on sustainable Genomics for national #AMR surveillance in Colombia, India, Nigeria, and the Philippines and local / global genomic context for ongoing surveillance of major #AMR pathogens. Here we detail our work with a series of preprints (1/10)
1
1
2