NingQing Liu
@NingQing_Liu
Followers
44
Following
9
Media
0
Statuses
24
A molecular biologist who studies gene regulation in normal and cancer cells, with special interests in 3D genome organization.
Erasmus MC
Joined December 2010
RT @bvansteensellab: New preprint by our lab -- Revealing the functional landscape of a large genomic locus by highly multiplexed local hop….
www.biorxiv.org
Genes are often activated by enhancers located at large genomic distances. The importance of this positioning is poorly understood. By relocating promoter-reporter constructs into >1,000 alternative...
0
22
0
RT @stefanogmanzo: One of my postdoctoral efforts from @bvansteensellab is now out on bioRxiv. We tried to tackle a longstanding question:….
0
5
0
RT @deWitLab: Our paper on patient-specific MYC enhancers in malignant rhabdoid tumors was just published in @NatureComms Please the link h….
www.nature.com
Nature Communications - The regulatory landscape of malignant rhabdoid tumor (MRT) due to SMARCB1 loss remains to be explored. Here, the authors perform multi-omics analysis using patient-derived...
0
14
0
RT @d_pastoors: Very excited to share our preprint (& my first (co)first-author preprint🤩) about targeting EVI1 in AML by disrupting its in….
0
16
0
RT @vanLeeuwenLab: Unlocking the mystery of tRNA gene regulation! Our latest pre-print reveals a new protein and new mechanism promoting RN….
0
13
0
RT @tinekelenstra: Excited that this work is now officially published! Using quantitative imaging of endogenous Gal4 in budding yeast, we s….
academic.oup.com
Abstract. Many transcription factors (TFs) localize in nuclear clusters of locally increased concentrations, but how TF clustering is regulated and how it
0
17
0
RT @MathijsSanders: What drives clonal expansion (CE) in humans? How do they relate to the developmental process of cancer? What sets their….
onlinelibrary.wiley.com
Our DNA is consistently assaulted by a variety of intrinsic and extrinsic mutational factors. Fortunately, DNA repair provides for protective barriers that limit the full manifestation of DNA...
0
22
0
RT @deWitLab: We are happy to present our new manuscript describing a role for long-range promoter-enhancer looping of the MYC gene in mali….
www.biorxiv.org
Malignant rhabdoid tumor (MRT) is a highly malignant and often lethal childhood cancer. MRTs are genetically defined by bi-allelic inactivating mutations in SMARCB1 , a member of the BRG1/BRM-assoc...
0
12
0
RT @biorxivpreprint: Identifying cross-lineage dependencies of cell-type specific regulators in gastruloids #bioRx….
0
1
0
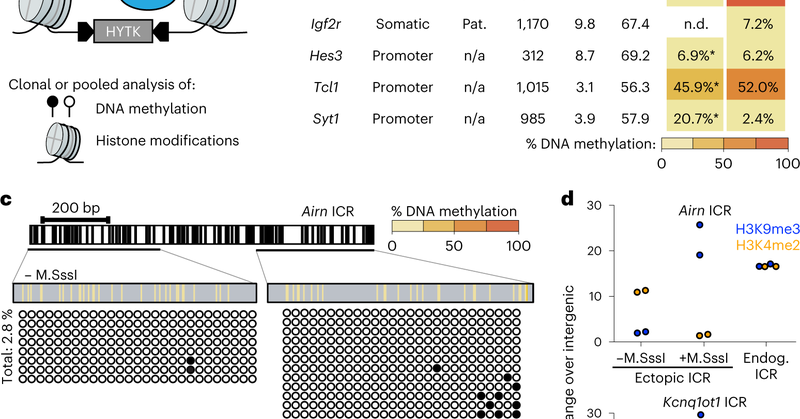
RT @TuncayBaubec: New publication spearheaded by former PhD student Stefan Butz published today. Systematic dissection of DNA sequence and….
www.nature.com
Nature Genetics - Ectopic imprinting control regions (ICRs) recapitulate chromatin states of endogenous imprinted loci in mouse embryonic stem cells. ATF7IP and ZMYM2 regulate epigenetic memory at...
0
68
0
Dynamic chromosomal interactions and control of heterochromatin positioning by Ki67
www.biorxiv.org
Ki67 forms a protective layer around mitotic chromosomes and is involved in genome positioning around nucleoli during interphase. However, a lack of genomic interaction maps has hampered a detailed...
0
0
1
Systematic analysis of intrinsic enhancer-promoter compatibility in the mouse genome
www.biorxiv.org
Gene expression is in part controlled by cis-regulatory elements (CREs) such as enhancers and repressive elements. Anecdotal evidence has indicated that a CRE and a promoter need to be biochemically...
0
0
3
RT @biorxivpreprint: CTCF and cohesin promote focal detachment of DNA from the nuclear lamina #bioRxiv.
0
11
0
RT @biorxivpreprint: Rapid depletion of CTCF and cohesin proteins reveals dynamic features of chromosome architecture. .
0
9
0
RT @AnnaGManjon: I am very excited to share our new manuscript on how Nuclear Lamina (NL) may regulate gene expression and promote drug res….
www.biorxiv.org
Acquired drug resistance is a major problem in the treatment of cancer. hTERT-immortalized, untransformed RPE-1 (RPE) cells can acquire resistance to taxol by derepressing the ABCB1 gene, encoding...
0
33
0
RT @deWitLab: Our WAPL degron story was just published in @NatureGenet. We find that both cohesin loading and off-loading is essential for….
www.nature.com
Nature Genetics - WAPL creates a pool of free cohesin that binds to cell-type-specific sites. This cohesin turnover is important for maintaining promoter–enhancer loops.
0
52
0
RT @deWitLab: This is published version of our bioRxiv pre-print. For a thread covering the main conclusions see:
0
1
0
RT @deWitLab: Biggest changes with the pre-print: 1) TT-seq to measure active transcription which really improves the number of differentia….
0
1
0