Jan-Niklas Tants
@JanNiklasTants
Followers
12
Following
8
Media
0
Statuses
28
Joined July 2023
Happy to see this work published! Check out how Roquin binding affects RNA structure; and some nice methods how to obtain such information. Thanks @lab_nmr and the Rief lab!
pnas.org
The interaction of mRNA and regulatory proteins is critical for posttranscriptional control. For proper function, these interactions, as well as th...
0
1
2
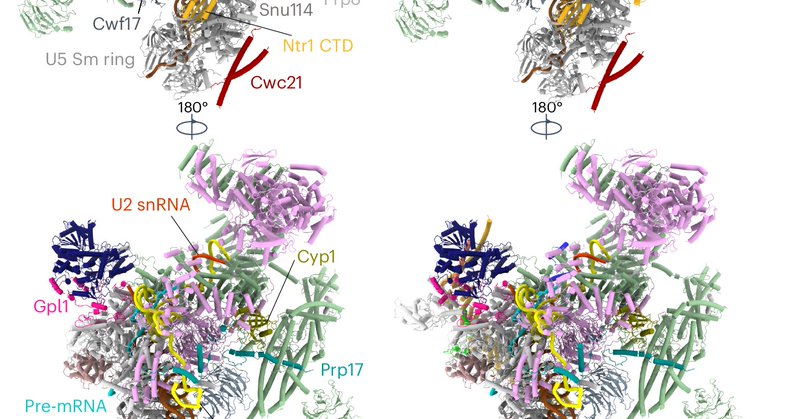
Spliceosomes use quality control mechanisms to detect and correct mistakes during RNA processing. Our work explores how defective spliceosomes are identified and discarded. #RNA #RNAProcessing #Spliceosomes #QualityControlMechanisms #RMAP
https://t.co/IjYgWUr7Pr
nature.com
Nature Structural & Molecular Biology - Here, the authors provide insights into a splicing quality control mechanism. The Gpl1–Gih35 complex binds to the active site of aberrant...
0
5
23
New preprint from @lab_nmr! We exploit a nice set of techniques to decipher the bimodal RNA-binding of Roquin. Thanks @andreas_walbrun and the Rief lab. https://t.co/Xjyk2Ol7CD
biorxiv.org
The interaction of mRNA and regulatory proteins is critical for post-transcriptional control. For proper function, these interactions as well as the involved protein and RNA structures are highly...
0
2
4
Very excited to share our latest preprint, led by rockstar post-doc @i_borovska. Our humble attempt to begin exploring #RNA secondary structure ensembles in a living cell ( https://t.co/XMHdvLLUq6) (1/n)
biorxiv.org
RNA molecules can populate ensembles of alternative structural conformations, but comprehensively mapping RNA conformational landscapes within living cells presents significant challenges and has, as...
7
14
77
Check out our new review @lab_nmr about RNA structure, associated diseases and the potential for RNA-targeted drugs! https://t.co/7ferrZmc7x
portlandpress.com
Abstract. Regulatory RNA elements fulfill functions such as translational regulation, control of transcript levels, and regulation of viral genome replication. Trans-acting factors (i.e., RNA-binding...
0
4
12
Nice collaboration on one of our most beloved proteins and its precise stem-looped RNA targets. Was a pleasure to contribute! Congrats to everyone @WeigandLab and @ david mathews lab! Have a look! 👇👇👇 https://t.co/IyqxKslfSG
onlinelibrary.wiley.com
sRBNS (structured RNA Bind-n-Seq) allows precise mapping of RNA-protein interactions including RNA structures at nucleotide resolution. Using this new method, we dissected the RNA-binding preferenc...
0
2
18
Evolution of the RNA alternative decay cis element into a high-affinity target for the immunomodulatory protein Roquin https://t.co/hBmrjpGr0t
#biorxiv_biochem
0
4
2
We are hiring! The Schlundt lab has an open postdoctoral position (2+ years) in #NMR -supported #RNA-protein structural biochemistry at beautiful @uni_greifswald! #biochemie #greifswald 👨🔬👩🔬 Interested? Then get in contact (see below)! Please RT and spread the word! 👇👇
0
26
38
Really happy to see this one finally out! Thanks @lab_nmr members for support and @WeigandLab for this collaboration! https://t.co/dEMdHA183y
academic.oup.com
Abstract. Zinc finger (ZnF) domains appear in a pool of structural contexts and despite their small size achieve varying target specificities, covering sin
0
2
10
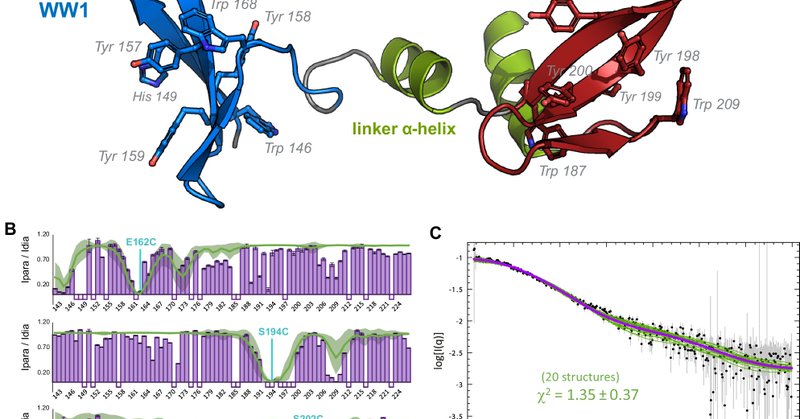
Check out our exciting new article from @Sattler_lab in collaboration with @koenig_lab published in @NatureComms: https://t.co/xBbBhNI1vd. We study the interaction between the human splicing factors PRPF40A and SF1 by #NMR, #SAXS and #ITC. We found…
nature.com
Nature Communications - The specific recognition of a proline-rich motif in the intrinsically disordered region of SF1 by the PRPF40A tandem WW domains is modulated by an intramolecular...
2
6
17
Another publication is online! 🎉 Biophysical Investigation of RNA•DNA:DNA triple helix and RNA:DNA heteroduplex formation by the lncRNAs MEG3 and Fendrr Can be found here: https://t.co/do937hcQ34
#lncRNA #NMR #smFRET #MST
chemistry-europe.onlinelibrary.wiley.com
It is known that lncRNAs can not only form RNA : DNA heteroduplexes, but above all RNA ⋅ DNA : DNA triplexes to perform regulatory functions. For a better understanding of the structure and functio...
0
5
11
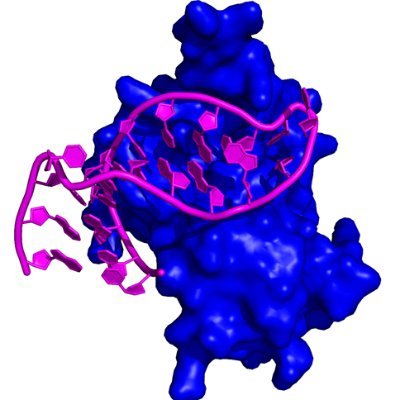
Structure and RNA-binding of the helically extended Roquin CCCH-type zinc finger https://t.co/my7BPRHbkq
#bioRxiv
0
1
1
PRC2 is reported to bind RNA. Yet, genetic experiments have raised serious questions about their functional relevance. Our paper revisits the biochemical evidence supporting widespread PRC2-RNA interactions and finds that many of these do not occur in vivo.
13
110
468
📢In recognition of his fundamental contributions to #ProteinStructure formation and the role of #chaperones, Prof. Johannes Buchner @buchnerlab_tum @TU_Muenchen will be awarded the #OttoWarburgMedal 2024 of the GBM and its sponsors @ElsevierConnect and @BBAjournals🏅Congrats🎉
5
21
91
Check out our newest publication in @NAR_Open 🥳 "RNA G-quadruplex folding is a multi-pathway process driven by conformational entropy" Full text here: https://t.co/7FbzOEUEvK
#quadruplex
academic.oup.com
Abstract. The kinetics of folding is crucial for the function of many regulatory RNAs including RNA G-quadruplexes (rG4s). Here, we characterize the foldin
0
2
14
New #KwokLab paper: Selective targeting of parallel G-quadruplex structure using L-RNA aptamer https://t.co/9o44klADEE Congrats to all the authors! @NAR_Open
@CityUScience
@CityUChem
@CityUHongKong
academic.oup.com
Abstract. G-quadruplexes (G4) are special nucleic acid structures with diverse conformational polymorphisms. Selective targeting of G-quadruplex conformati
1
5
27
AlphaFold, small-angle X-ray scattering and ensemble modelling: a winning combination for intrinsically disordered proteins
1
7
22
Pleased to share our new article "In vivo-like nearest neighbor parameters improve prediction of fractional RNA base-pairing in cells" out in Nucleic Acids Research. Collaboration with @BabitzkePaul lab. https://t.co/QqOLobj3Sj
3
25
113
👋Prof. Michael Sattler welcomes ca. 150 participants from many countries to celebrate the happy end of a 10 year long journey: the 1.2 Gigaherz #Spectrometer (built by @bruker, funded by #Wissenschaftsrat and #DFG). It is up and running and provides great insight into
2
4
33
"Atomistic structure of the SARS-CoV-2 pseudoknot in solution from SAXS-driven molecular dynamics" — Read it on @ResearchGate:
researchgate.net
PDF | SARS-CoV-2 depends on −1 programmed ribosomal frameshifting (−1 PRF) to express proteins essential for its replication. The RNA pseudoknot... | Find, read and cite all the research you need on...
0
1
1