@HannesStaerk
@BarzilayRegina
The project resulted from a great collaboration with
@HannesStaerk
(joint first), Bowen Jing (joint first),
@BarzilayRegina
and Tommi Jaakkola. Code and trained models are available at:
2
0
22
Replies
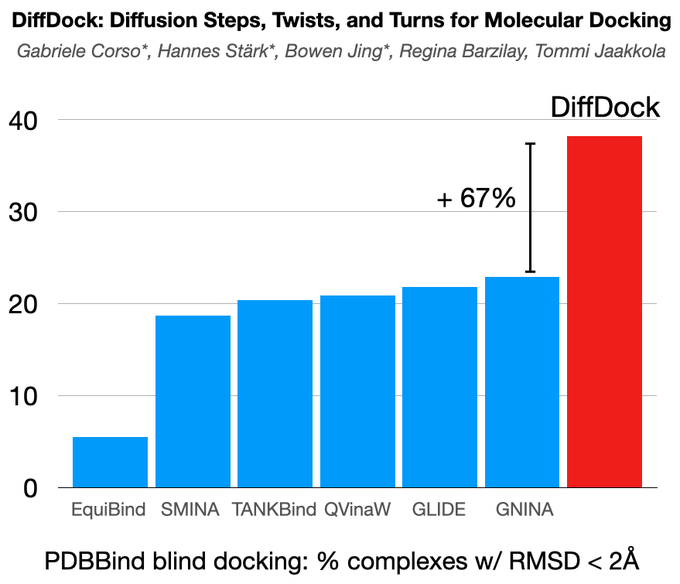
Excited to share DiffDock, new non-Euclidean diffusion for molecular docking! In PDBBind, standard benchmark, DiffDock outperforms by a huge margin (38% vs 23%) the previous state-of-the-art methods that were based on expensive search!
A thread! 👇
16
96
450
@HannesStaerk
@BarzilayRegina
Recent regression-based ML methods for docking showed strong speed-up but no significant accuracy improvements over traditional search-based approaches. We identify the problem in their objective functions and show how generative modeling aligns well with the docking task.
2
1
14
@HannesStaerk
@BarzilayRegina
We, therefore, develop DiffDock, a non-Euclidean diffusion model over the space of ligand poses for molecular docking. DiffDock defines a diffusion process over the position of the ligand relative to the protein, its orientation, and the torsion angles describing its conformation
1
1
11
@HannesStaerk
@BarzilayRegina
To efficiently train and run the diffusion model over this highly non-linear manifold, we map the elements of the manifold to a product space of T(3) x SO(3) x SO(2)^m groups corresponding to the translation, rotation, and torsion transformations.
1
0
6
@HannesStaerk
@BarzilayRegina
We achieve a new state-of-the-art 38% top-1 prediction with RMSD<2A on the PDBBind blind docking benchmark, considerably surpassing the previous best search-based (23%) and deep learning methods (20%). DiffDock also has faster runtimes than the previous state-of-the-art methods.
1
0
10
@HannesStaerk
@BarzilayRegina
We also train a confidence model that can rank poses by likelihood and predict the model's confidence in the generated poses. Experimentally, this confidence score provides high selective accuracy, reaching 83% on its most confident third of the previously unseen complexes.
1
0
7
1
1
13
@GabriCorso
@HannesStaerk
@BarzilayRegina
👏 Excellent work! 👏 Congrats!! 🎉
Have you looked at how it compares to bounding box docking?
1
0
1
@CyrusMaher
@HannesStaerk
@BarzilayRegina
Thank you! We have not evaluated our method when using a bounding box, however, we have compared it to the common approach of using a pocket prediction method (like P2Rank or EquiBind) followed by bounding box docking using search-based methods.
1
0
3