Angelique Deleris
@AtDeleris
Followers
153
Following
178
Media
0
Statuses
46
Joined August 2017
Our new study on characterizing plant DNA methylation reader proteins, led by @jon_cahn and @JamesPBLloyd, and great collaboration with the Michiel Vermeulen and @ahmillar9 groups
0
15
43
Our long standing review on T-DNA epigenetic regulation during plant transgenesis and pathogenesis is finally out ! Congrats @joaquinfrp for all the work reviewing the littérature since 1982! Such a fascinating mobile element and complex regulation!
academic.oup.com
A review of the mechanisms that silence the T-DNA in transgenic plants and during Agrobacterium infection and the crosstalk between the T-DNA and genome-wi
1
23
82
very happy to present our latest work on the diversity of RiboSeq signals associated with canonical and noncanonical translation! An almost 100% @i2bc paper with @BioCgpapado and @namyolivier published in @GenomeBiology ! https://t.co/xGkK7wbxUm
link.springer.com
Genome Biology - Pervasive translation is a widespread phenomenon that plays a critical role in the emergence of novel microproteins, but the diversity of translation patterns contributing to their...
4
8
24
Very happy and proud to share the preprint of our integrative study revealing a unique mechanistic framework for a chromatin switch mediated by a dual trxG/PcG factor. Made possible thanks to the work and dedication of several PhD students & postdocs, and fruitful collaborations.
The dual trxG/PcG protein ULTRAPETALA1 modulates H3K27me3 and directly enhances POLYCOMB REPRESSIVE COMPLEX 2 activity for ... https://t.co/dESO4gf70x
#biorxiv_genetic
2
20
44
Spécial thanks and congrats to Valentin Hure for his great work and thanks ANRJCJC for funding that was fruitful!
1
0
3
Together, our results uncover a novel crosstalk between DNA methylation and Polycomb at TEs and reveal that these pathways, thought to be specialized and antagonistic, can be interdependent and cooperate more than anticipated to maintain genome/epigenome integrity in eukaryotes.
0
0
3
3) Our work further reveals a new function of active DNA demethylation in modulating H3K27me3 levels in vegetative tissues, which we confirmed via targeted DNA methylation experiments.
1
0
1
2) Accordingly, H3K27me3 can be promoted by DNA methylation at some endogenous TEs while it is antagonized by DNA methylation at others (dual, locus-specific effect)
1
0
1
1) The de novo DNA methyltransferase DRM2 is unexpectedly required for H3K27me3 deposition at a neo-inserted Transposable element (TE) sequence
1
0
3
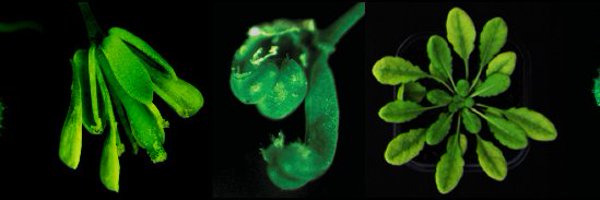
Very proud to share our second pre-print of the year, this time on the positive crosstalk between DNA methylation and Polycomb-group proteins at Arabidopsis transposable elements (TEs): https://t.co/YVtJWeOaPG
biorxiv.org
Transposable elements are primarily silenced by DNA methylation and the associated histone modification H3K9me2 in many multicellular eukaryotes, including plants. However, in the absence of DNA...
8
38
130
An atlas of conserved transcription factor binding sites reveals the cell type-resolved gene regulatory landscape of flowering plants https://t.co/jnbYw5K0wQ
#biorxiv_genomic
1
31
64
So proud of my lab for our latest work out today in @Naturepub. We took advantage of transposable elements and CRISPR to perform targeted insertion of cargo DNA in the Arabidopsis and soy genomes. Led by @mcliupeng. https://t.co/wSvr1uuEbF. 🧵
nature.com
Nature - Fusion of rice Pong transposase to the Cas9 or Cas12a programmable nucleases provides sequence-specific targeted insertion of enhancer elements, an open reading frame and gene expression...
29
157
436
Last week to register for the @epiplant / @SEBiology symposium on Plant Epigenetics, @GReD_Clermont Clermont-Ferrand, France, July 10-12, 2024. Program & registration ➡️ https://t.co/1vF7GNiJpe Deadline 30th of April!
0
5
16
Hello world! We are looking to hire : - a Master 2 student (with possibility for a PhD) contact: chloe.girard [a] https://t.co/xw0t55pS4A - Lab engineer (molecular biology and microscopy) apply here: https://t.co/k04EvfV8eW Details on the project: https://t.co/5BByRQ3wDJ
3
78
95
The last 100% home made story about (epi)genome surveillance interplays 🌱🧬☀️…Enjoy !!!!! The histone demethylase JMJ27 acts during the UV-induced modulation of H3K9me2 landscape and facilitates photodamage repair | bioRxiv
biorxiv.org
Plants have evolved sophisticated DNA repair mechanisms to cope with the deleterious effects of UV-induced DNA damage. Indeed, DNA repair pathways cooperate with epigenetic-related processes to...
1
8
19
💥Our new preprint! Transposon ADDICTION in development! Gag proteins of endogenous retroviruses are required for zebrafish development Study led heroically by @sylviaandbu & @JN_Wells This work has transformed my view of transposons, no less! 🧵1/n https://t.co/d1yNgFbBSb
biorxiv.org
Transposable elements (TEs) make up the bulk of eukaryotic genomes and examples abound of TE-derived sequences repurposed for organismal function. The process by which TEs become coopted remains...
20
101
342
In sum, we document an alternative control of TEs by H3K27me3 which is more plastic than DNA methylation, and that a TE can change epigenetic silencing state throughout its lifespan. This challenges the dogma that Arabidopsis TEs are solely targeted by DNA methylation in nature.
1
0
7
We show i) that some TEs are targeted by H3K27me3 (Polycomb) instead of DNA methylation in wildtype Arabidopsis ii) H3K27me3 can be deposited on newly inserted TE sequences iii) the existence of “bifrons” TEs that are marked by DNA methylation or H3K27me3 depending on the ecotype
1
0
6